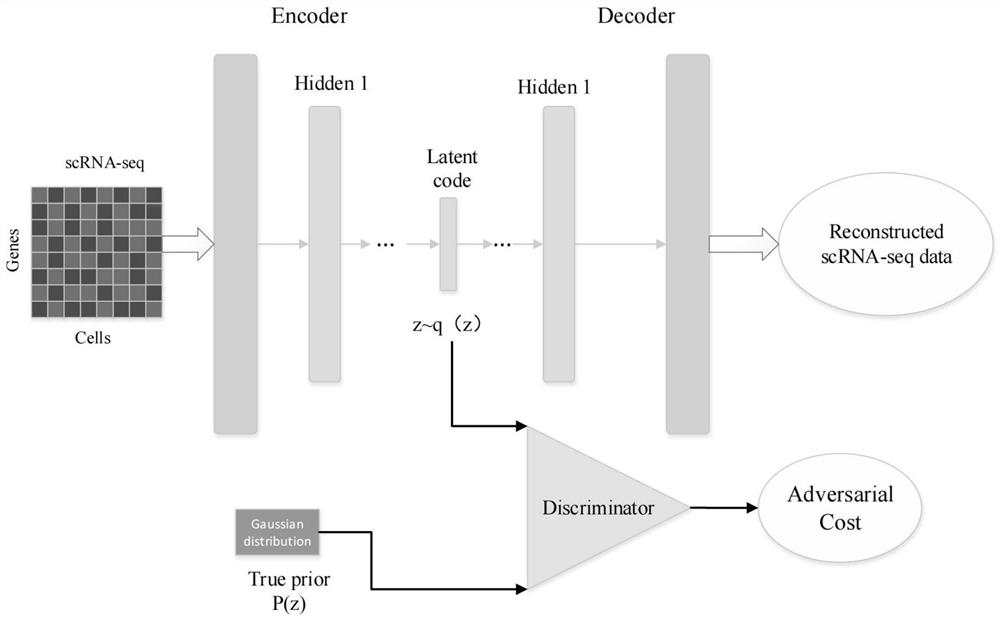
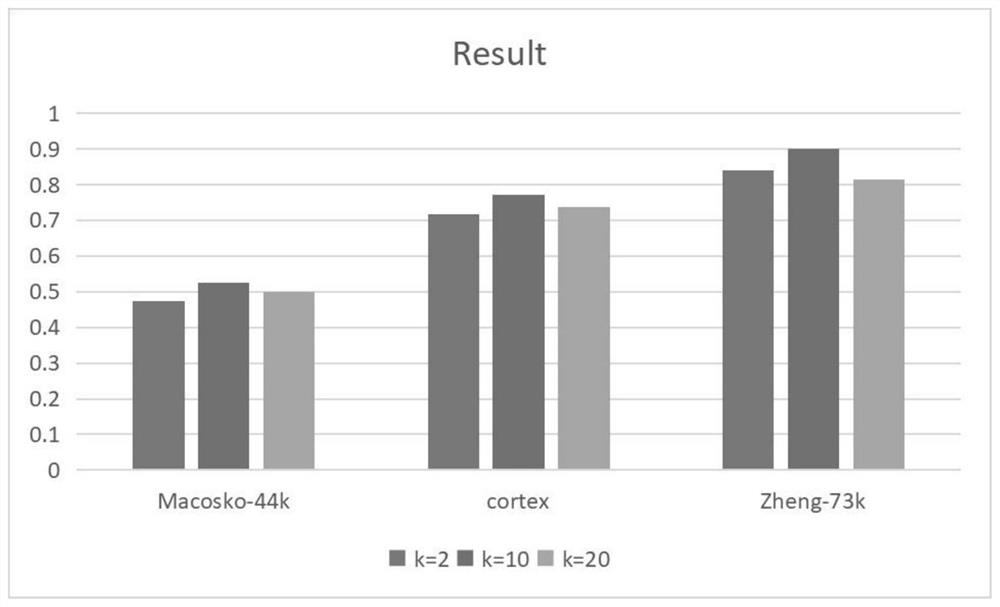
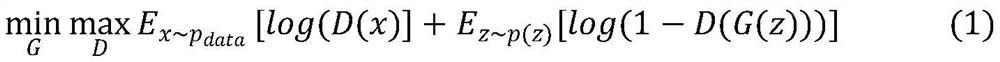ScRNA-seq data dimension reduction method based on deep adversarial variational auto-encoder
An autoencoder and data dimensionality reduction technology, applied in neural learning methods, instruments, biological neural network models, etc., can solve problems such as loss of important information and insufficient feature extraction.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0054] In order to make the object, technical solution and advantages of the present invention clearer, the present invention will be further described in detail in combination with experiments below. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
[0055] 1. Dataset overview
[0056] We evaluate the proposed SCAVAE model on three real scRNA-seq datasets from different sequencing platforms. All the datasets used in this paper are publicly available, and the statistics of the datasets are summarized in Table 1. In each dataset, 720 genes with the largest variance were selected for subsequent experiments. Details are shown in Table 1:
[0057] Table 1 Datasets used in this experiment
[0058]
[0059] 2. Experimental environment and parameter settings
[0060] The hardware environment is mainly a PC host. Among them, the CPU of the PC host is 11th Gen Intel(R) Core(T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com