Acid sphingomyelinase gene and diagnosis of Niemann-pick disease
a technology of acid sphingomyelinase and gene, applied in the field of acid sphingomyelinase gene and methods of diagnosing niemannpick disease, can solve the problems of inability to correlate disease severity, inability to reliably distinguish two phenotypes in biochemical analysis, and difficulty in enzymatic detection of obligate heterozygotes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0021] For purposes of clarity, and not by way of limitation, the detailed description of the invention is divided into the following subsections:
[0022] (i) the ASM gene;
[0023] (ii) expression of the ASM gene;
[0024] (iii) identification of mutations in the ASM gene;
[0025] (iv) methods of diagnosing Niemann-Pick disease;
[0026] (v) assay systems for diagnosing Niemann-Pick disease; and
[0027] (vi) methods of treatment of Niemann-Pick disease.
5.1. The Acid Sphingomyelinase Gene
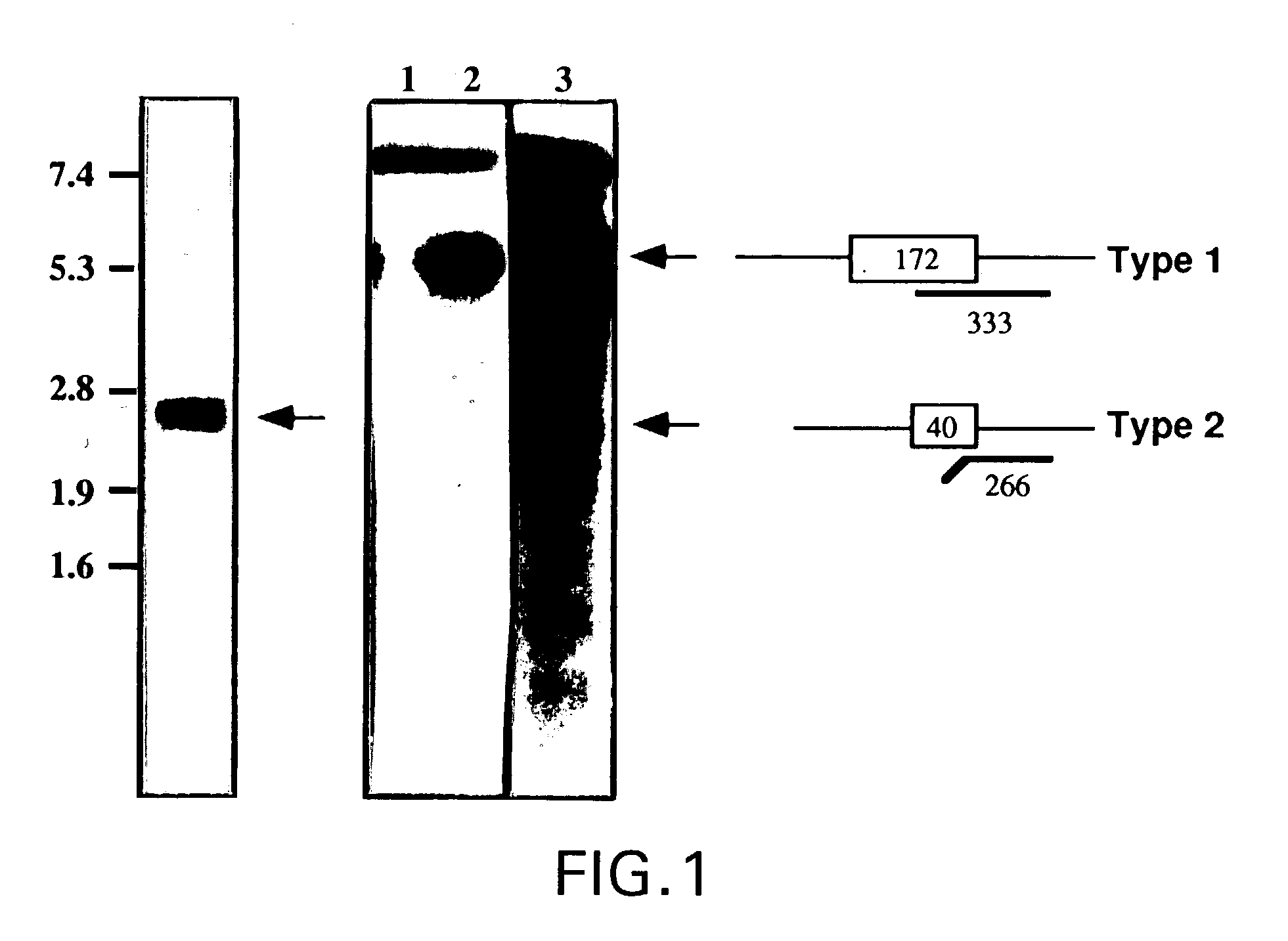
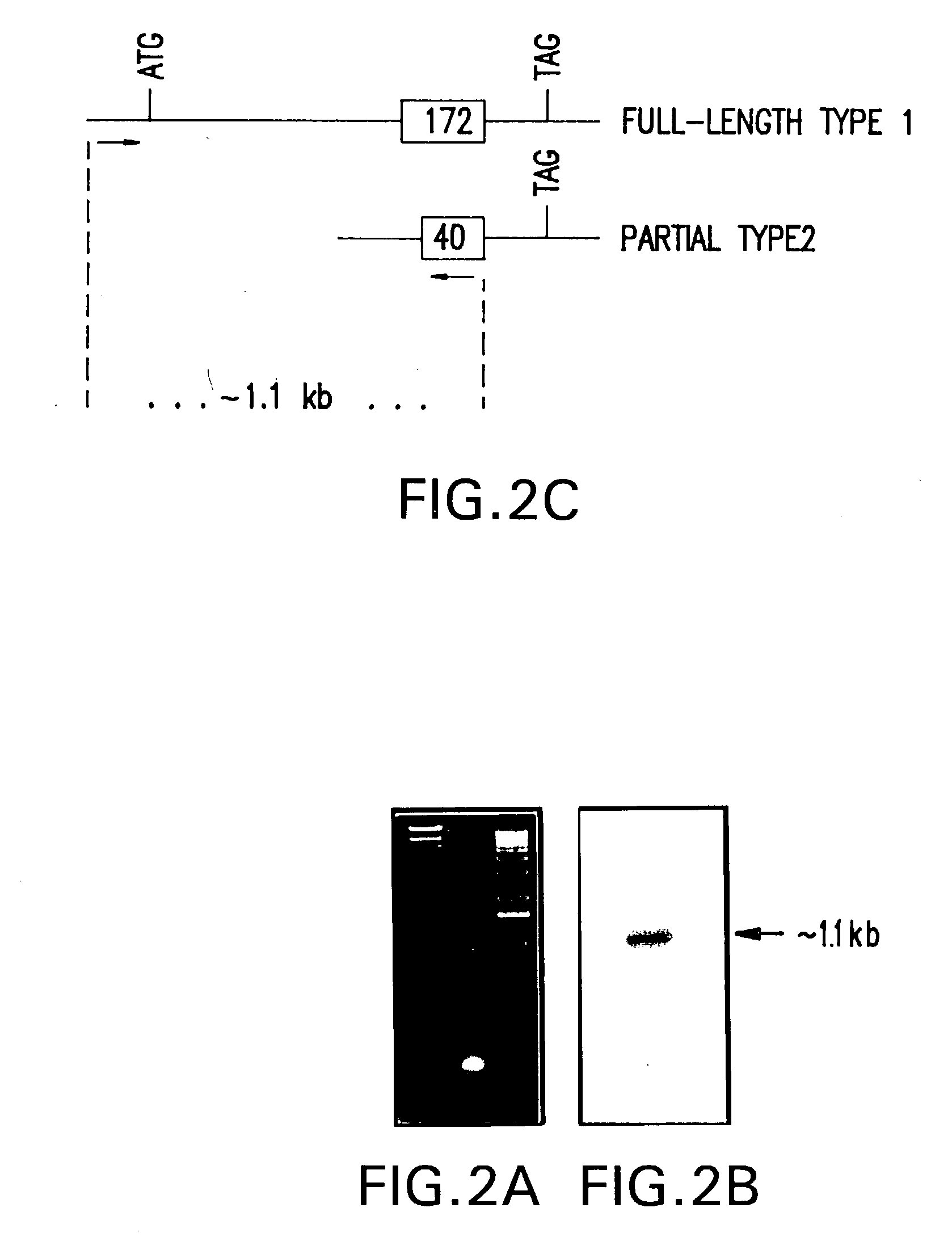
[0028] The nucleotide coding sequence and deduced amino acid sequence for the full-length cDNA that encodes functional ASM is depicted in FIG. 3, and is contained in plasmid pASM-1FL, as deposited with the NRRL and assigned accession no. ______. This nucleotide sequence, or fragments or functional equivalents thereof, may be used to generate recombinant DNA molecules that direct the expression of the enzyme product, or functionally active peptides or functional equivalents thereof, in appropriate host cells. The g...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com