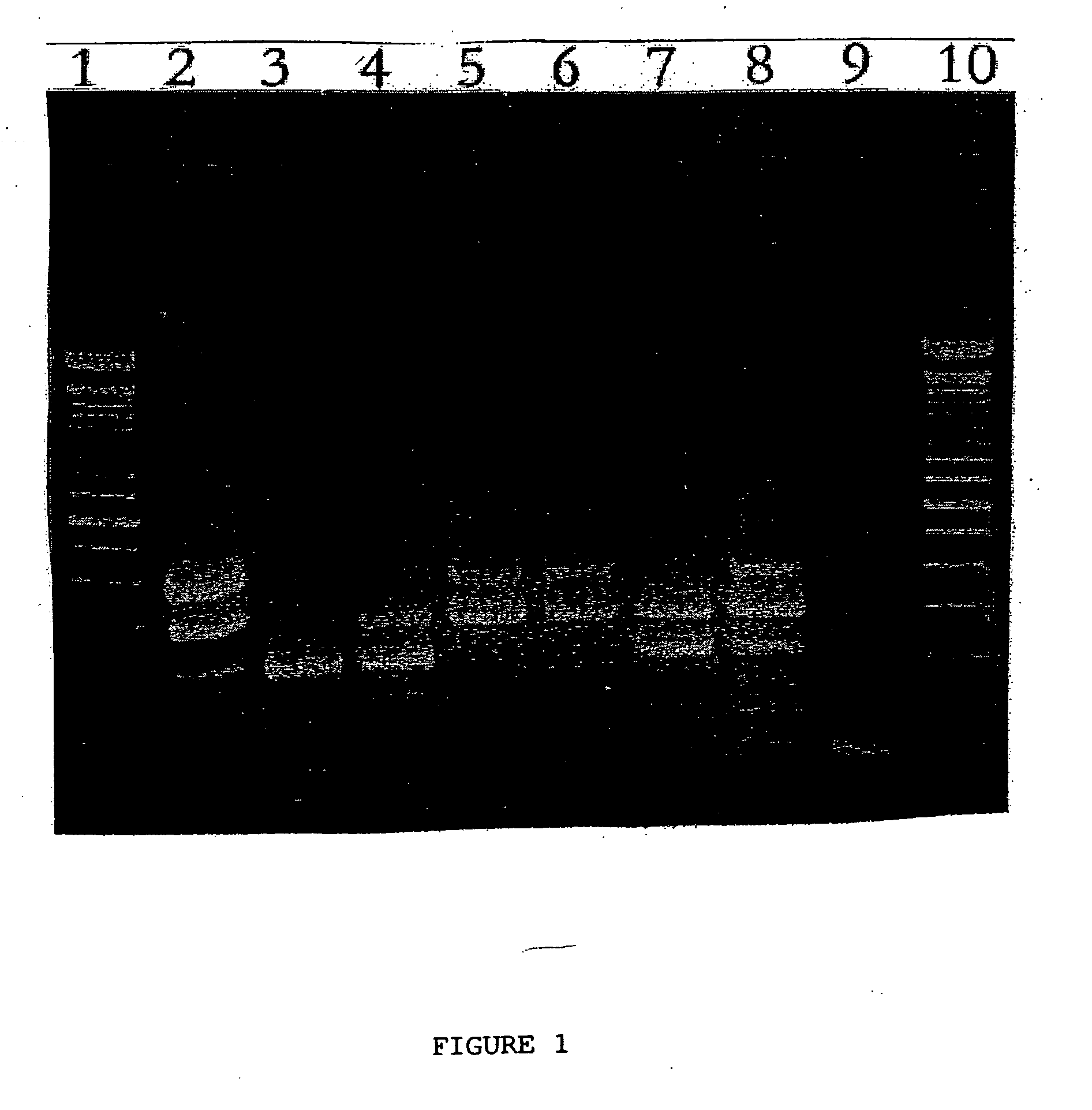
Multiplex RT-PCR/PCR for simultaneous detection of bovine coronavirus, bovine rotavirus, Cryptosporidium parvum, and Escherichia coli
a multi-core, rtpcr technology, applied in the field of multi-core rtpcr/pcr method, can solve the problems of high cost, time-consuming and laborious process for detecting these pathogens, and currently used assays have severe limitations in detecting these agents, etc., to achieve rapid detection, sensitive, and specific
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0064] This example shows a method for recovering nucleic acids from feces. Both RNA and DNA were simultaneously extracted from 12 fecal specimens from calves with diarrhea submitted to the Animal Health Diagnostic Laboratory at Michigan State University using the Qiagen DNEASY Tissue Kit following the DNEASY protocol for animal tissues in the DNEASY Tissue Kit Handbook (Qiagen, Inc., April 1999 Edition, pp. 16-18) provided by the manufacturer as modified below.
[0065] About 20 to 30 mg of feces were resuspended in 180 μl ATL buffer (Qiagen, Inc.). To lyse the cells in the suspension, 20 μl of proteinase K was added and the suspension was incubated for 30 minutes at 55° C. Afterwards, to disrupt the C. parvum oocysts, four freeze-thaw cycles were performed. Each freeze-thaw cycle consisted of freezing the suspension in a dry ice-acetone bath for 2 minutes followed by thawing in a 37° C. waterbath for another 2 minutes. After the last freeze-thaw, the suspension was centrifuged at 15...
example 2
[0067] This example shows the multiplex RT-PCR / PCR assay of the present invention using the nucleic acids isolated in Example 1 to detect bovine corona virus, bovine rotavirus, and Cryptosporidium parvum.
[0068] A single reaction tube was used for the multiplex reaction of each nucleic acid sample from Example 1. In this example, 12 separate multiplex reactions were performed.
[0069] The multiplex reactions were performed using the Qiagen ONESTEP RT-PCR Kit in a final reaction volume of 50 μl with the following final primer concentrations. The amount of each primer was as follows: 0.6 μM of each Cryptosporidium parvum primer (SEQ ID NO:1 and SEQ ID NO:2); 0.9 μM of each bovine coronavirus primer (SEQ ID NO:3 and SEQ ID NO:4); and, 0.6 μM of each bovine rotavirus primer (SEQ ID NO:5 and SEQ ID NO:6).
[0070] Prior to performing the PCR step of the reaction, i.e., before adding the enzyme mix, the samples containing the nucleic acids were exposed to a denaturing step to denature the do...
example 3
[0076] This example illustrates separation of bovine coronavirus, bovine rotavirus, Cryptosporidium parvum, and Escherichia coli K99 from a fecal sample.
[0077] Dynabeads pan mouse IgG (Dynal, Inc. #110.21) are resuspended thoroughly in their storage vial. Four tubes are set up and to each of the tubes, 25 μl of 107 beads per ml is transferred. The tubes are placed in a magnetic block (Home Depot) and the fluid is removed. The beads are washed twice with washing buffer (phosphate buffered saline (PBS) containing 1% bovine serum albumen (BSA)). Next, for each tube, the beads are incubated with mouse monoclonal antibodies against one of bovine coronavirus, bovine rotavirus, Cryptosporidium parvum, or Escherichia coli K99 in washing buffer at 1 μg of antibodies for every 107 beads. Thus, four tubes are prepared, each containing antibodies against one of the four pathogens. The antibodies are bound to the IgG on the beads by incubating for 2 hours at 4° C. with gentle mixing using a mix...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com