Affinity sensor for detecting specific molecular binding events and use thereof
a technology of molecular binding events and affinity sensors, applied in the field ofaffinity sensors, can solve the problems of inability to detect, low sensitivity and spatial resolution of signals emitted by individual marker molecules, and the limitations of the current automation of image analysis on the basis of chip technology, and achieve the effect of low expenditur
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0031] The invention will be explained hereinafter in more detail by virtue of schematical embodiments under reference to the drawings. There is shown in:
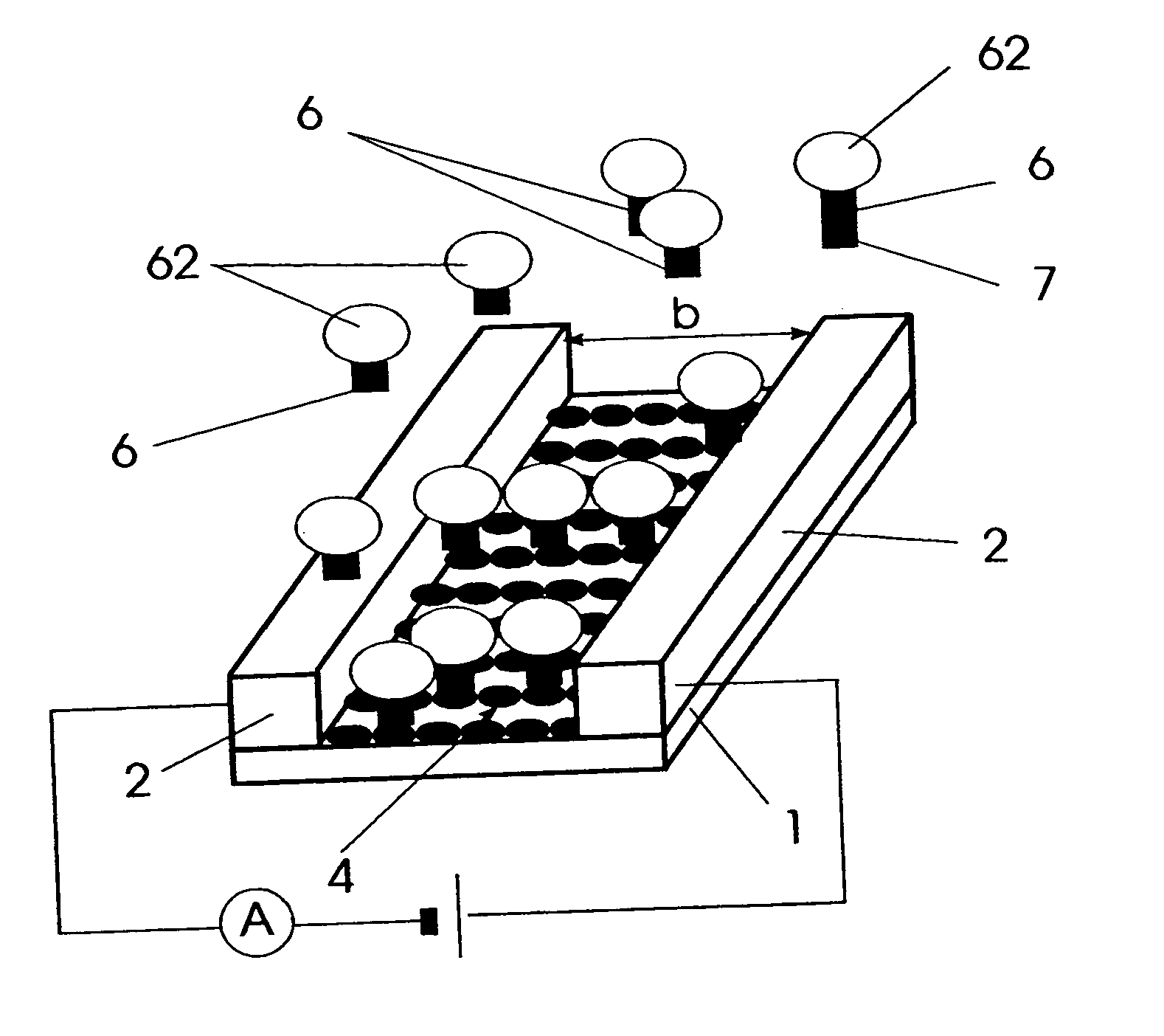
[0032]FIG. 1 an affinity sensor for detecting specific molecular binding events,
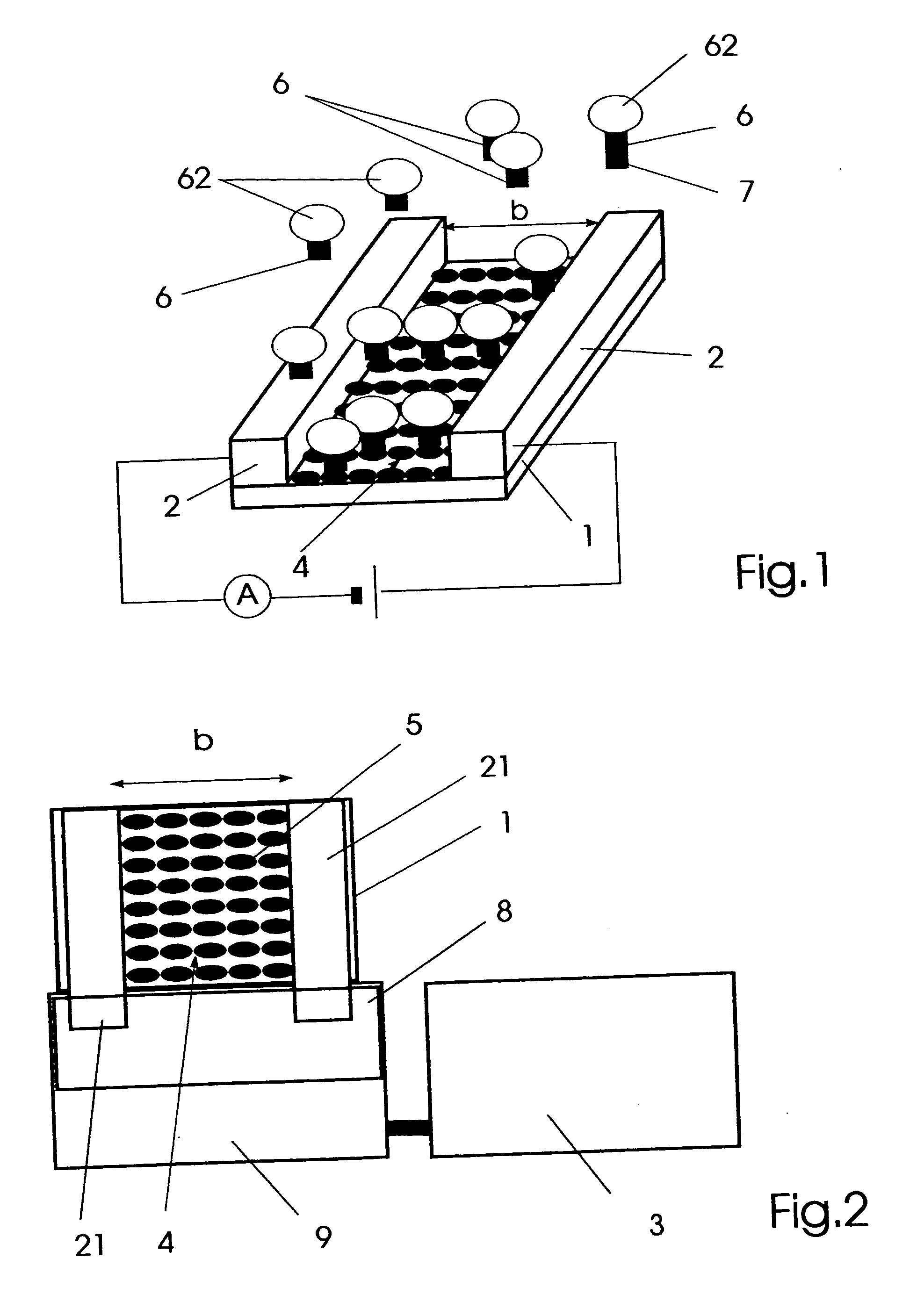
[0033]FIG. 2 a schematical representation of the affinity sensor for detecting specific molecular binding events,
[0034]FIG. 3 a cross-sectional view of an embodiment of the affinity sensor for detecting specific molecular binding events,
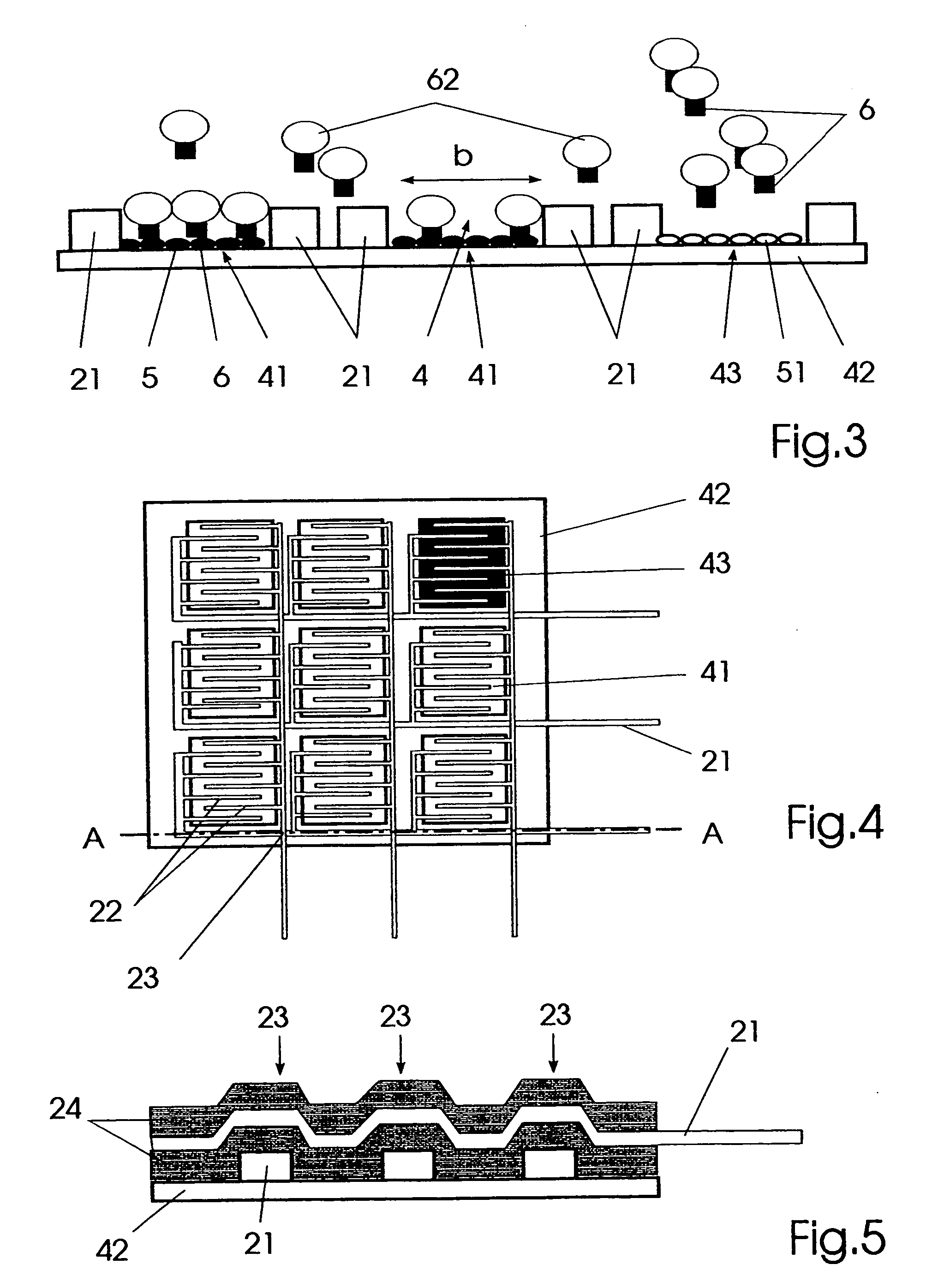
[0035]FIG. 4 a plan view of an embodiment of the affinity sensor in the form of an affinity chip, and
[0036]FIG. 5 a sectional view along the plane A-A of the affinity chip represented in FIG. 4.
DETAILED DESCRIPTION
[0037] An affinity sensor for detecting specific molecular binding events shown in FIGS. 1 and 2, is comprised of a carrier substrate 1 which is provided with electrodes 2 enclosing an area 4 that is provided with immobilized specific binding partners 5. Thereby the area 4 represents a discontinuity in an...
PUM
| Property | Measurement | Unit |
|---|---|---|
| height | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com