Compositions, kits, and methods for identification, assessment, prevention, and therapy of cervical cancer
a technology for cervical cancer and kits, applied in the field of cervical cancer, can solve the problems of difficult to achieve the effect of reducing the expression level of the marker, no absolute success guarantee, and more serious clinical problems,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
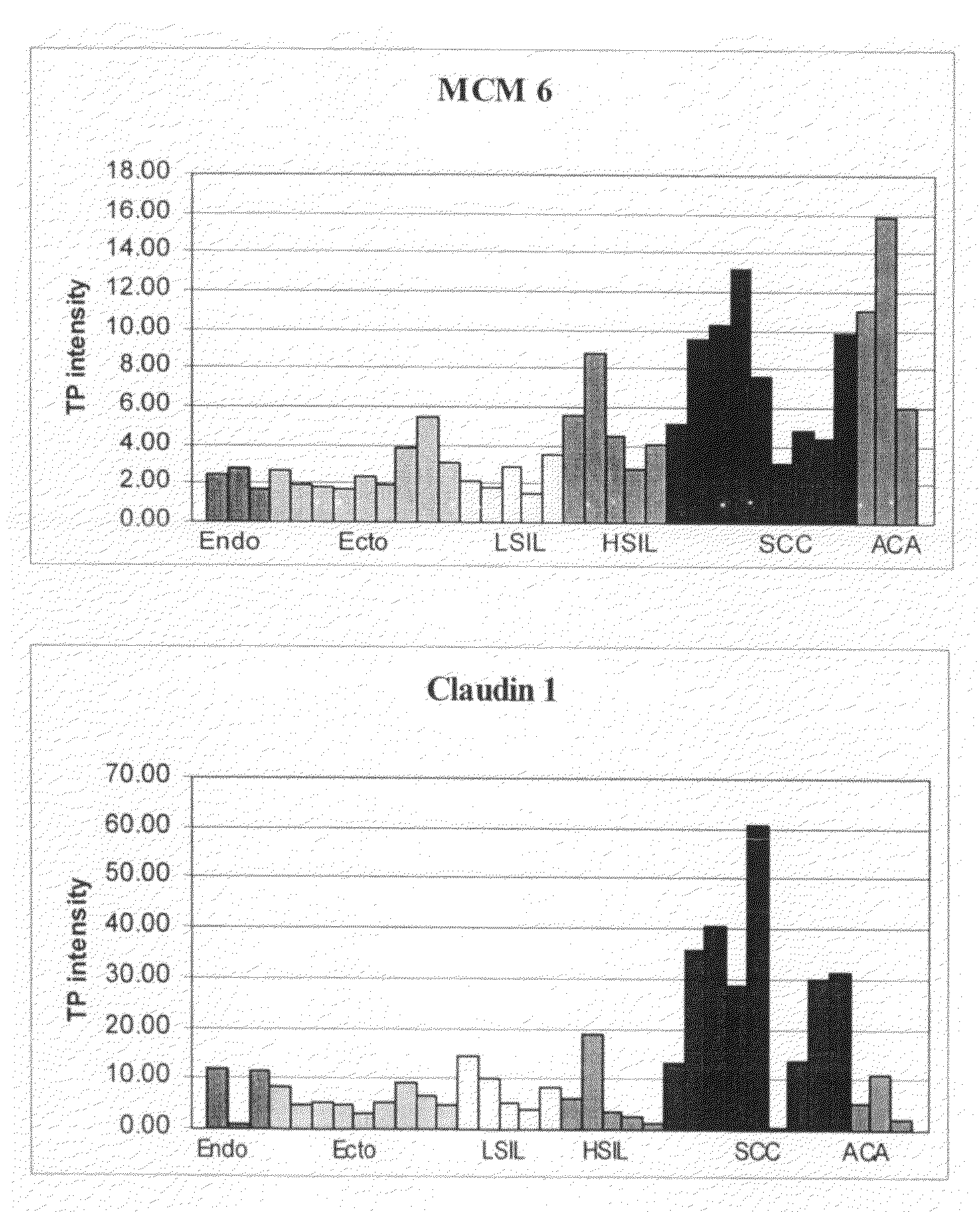
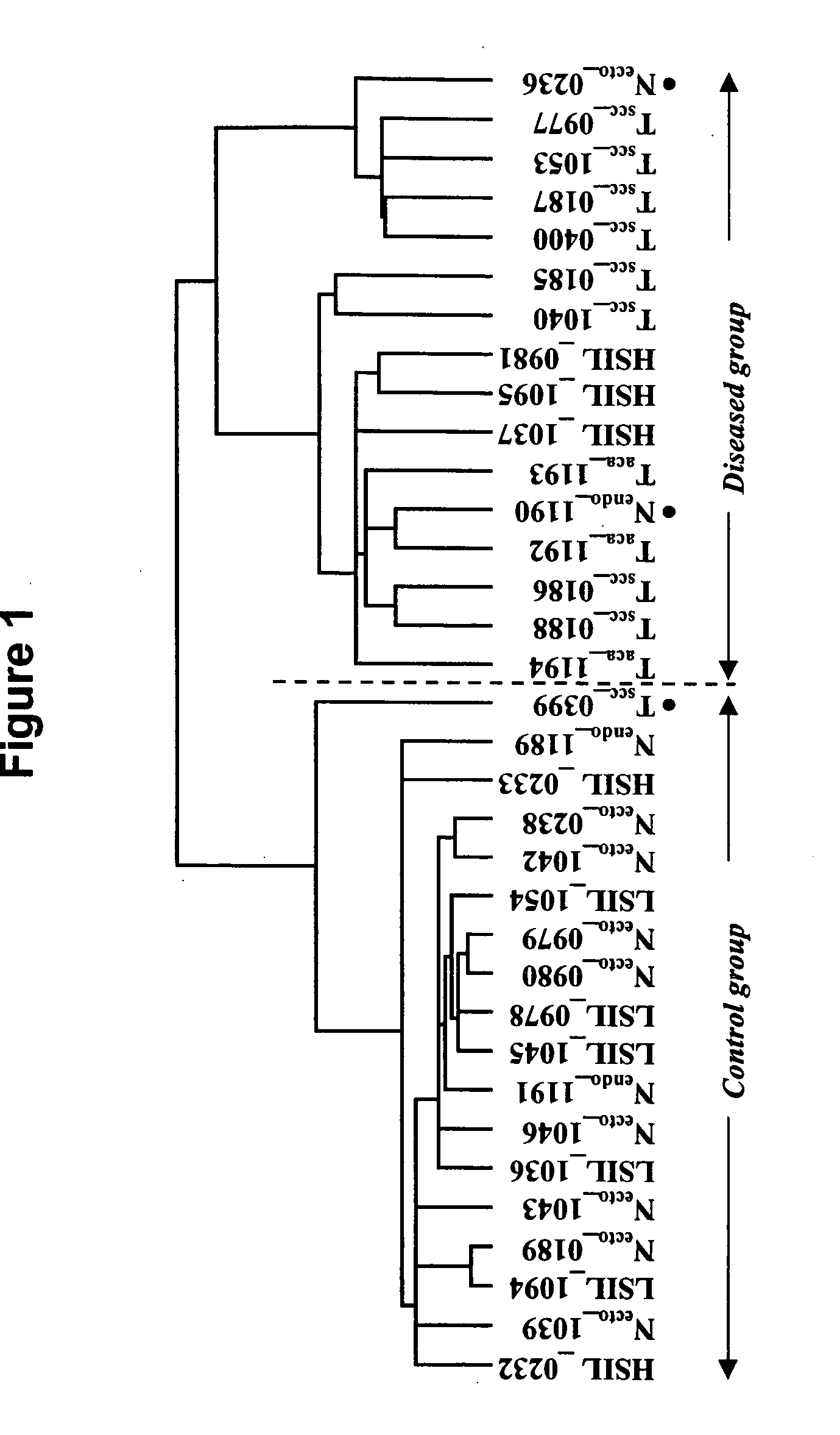
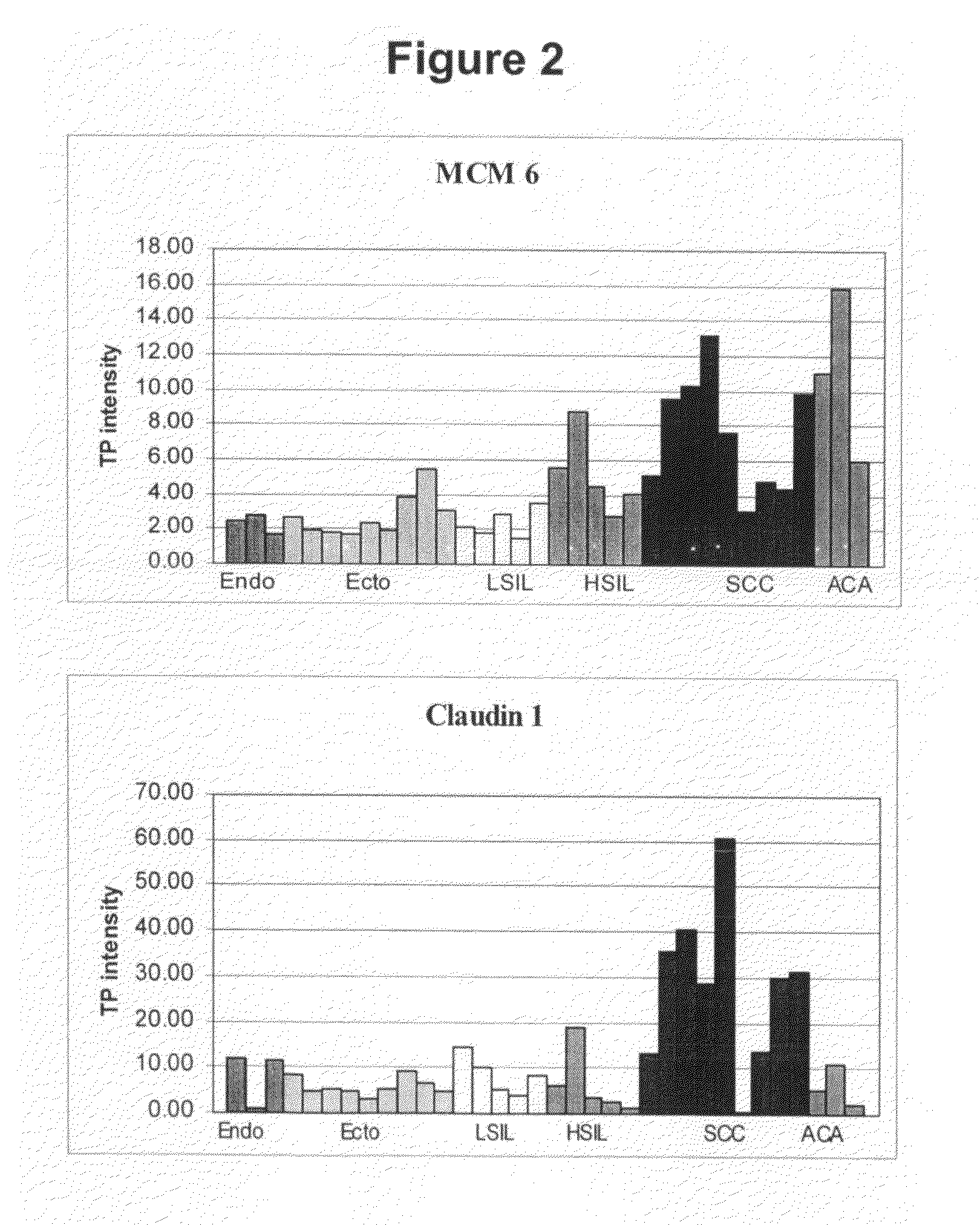
Identification of Cervical Cancer Markers by cDNA and Tissue Microarrays
I. Materials and Methods
Sample Collection and RNA Preparation
[0310]Cervical tissues were collected and snap frozen in liquid nitrogen. The histology and cellular composition of tissues were confirmed before RNA extraction was performed. Total RNA was extracted from the frozen tissues using Trizol Reagent (Life Technologies) followed by a secondary clean up step with Qiagen's RNeasy kit to increase RNA probe labeling efficiency (Qiagen, Valencia Calif.). Only RNA with a 28S / 18S ribosomal RNA ratio of at least 1.0, calculated using Agilent Technologies 2100 Bioanalyzer (Palo Alto, Calif.), was used in this study.
cDNA Microarray Hybridization
[0311]cDNA microarrays containing 30,732 Unigene clones from Research Genetics (Hunstville, Ala.) were generated on nylon filters. A total of 4-6 ug of total RNA was used as template to generate radioactively labeled cDNA by reverse transcription with 33P-dCTP, oligo dT-30 prim...
example 2
Gene Expression Analysis
RNA Preparation
[0328]Total RNA was prepared from various human tissues by a single step extraction method using TRIZOL Reagent according to the manufacturer's instructions (Invitrogen). Each RNA preparation was treated with DNase I (Ambion) at 37° C. for 1 hour. DNAse I treatment was determined to be complete if the sample required at least 38 PCR amplification cycles to reach a threshold level of fluorescence using β-2 microglobulin as an internal amplicon reference (or 35 PCR amplification cycles for 18s ribosome gene). The integrity of the RNA samples following DNase I treatment was confirmed by agarose gel electrophoresis and ethidium bromide staining. After phenol extraction, cDNA was prepared from the sample using the Taqman Reverse Transcription Reagents following the manufacturer's instructions (Applied Biosytems). A negative control of RNA without reverse transcriptase was mock reverse transcribed for each RNA sample.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com