Novel endoribonuclease
A technology of endoribonucleic acid and enzymatic activity, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Example 1: Isolate PH1182 from Pyrococcus Horikoshii ATCC700860, and construct an expression plasmid nucleotide sequence. Primer PH1182-F (SEQ ID NO: 3) and primer PH1182-R (SEQ ID NO: 4) for PCR amplification of the DNA region encoding the complete polypeptide were synthesized based on the nucleotide sequence information of PH1182.
[0069] Pyrococcus horikoshii ATCC700860 genomic DNA was obtained from ATCC (ATCC No. 700860D). PCR was performed with Pyrobest DNA polymerase (Takara Bio) and 50 ng of genomic DNA from Pyrococcus horikoshii ATCC700860 and primers PH1182-F and PH1182-R to obtain a 437-bp amplified DNA fragment. This amplified fragment was digested with restriction enzymes NdeI and XhoI, and subjected to agarose gel electrophoresis, and a 416-bp DNA fragment was recovered from the gel.
[0070] Expression vectors were constructed using pCold TF (Takara Bio). In order to introduce a stop codon after gene cloning following the XhoI site at the 3' end of the...
Embodiment 2
[0072] Example 2: Preparation of PH1182 polypeptide derived from Pyrococcus Horikoshii ATCC700860
[0073] Escherichia coli BL21(DE3) (Novagen) was transformed with the expression vector pCold TF-PH1182 obtained in Example 1 to obtain Escherichia coli pCold TF-PH1182 / BL21(DE3) for expression. Escherichia coli cells were cultured at 37°C in 5 ml of LB medium containing 100 µg / ml ampicillin. When OD600nm reached 0.5, incubate at 15°C for 30 minutes, add IPTG (Takara Bio) at a final concentration of 1 mM to induce polypeptide expression, and continue culturing at 15°C for 24 hours. The culture was terminated after 24 hours, and the cells were collected by centrifugation. Cells were suspended in 300 μl lysis buffer (50 mM NaH 2 PO 4 , 300 mM NaCl, 10 mM imidazole, pH 8.0), and disrupt the cells with a sonicator (Handy sonic, Tomy). To the supernatant collected by centrifugation was added 20 μl of Ni-NTA agarose (Qiagen), and the mixture was allowed to stand at 4° C. for 30 min...
Embodiment 3
[0074] Example 3: Using oligoribonucleotides as a substrate to identify the nucleotide sequence specificity of the PH1182 polypeptide
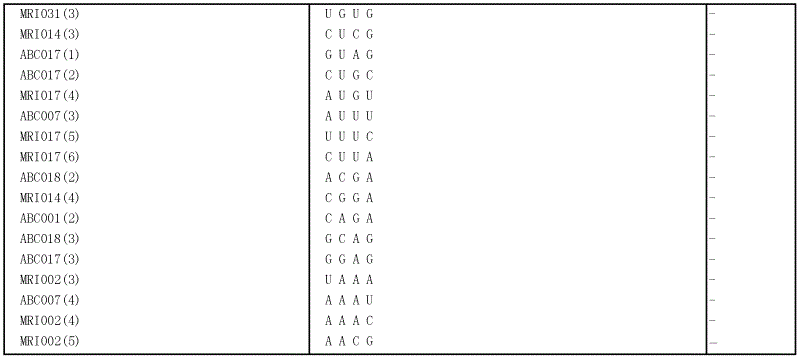
[0075] Oligoribonucleotides were synthesized and cleavage assays were performed to investigate the nucleotide sequence specificity of the ribonuclease activity of the PH1182 polypeptide obtained from Example 2.
[0076] Eleven oligoribonucleotides of SEQ ID NOS: 7-17 were synthesized as substrates. A 5-µl reaction mixture consisting of 10 µM of one of the oligoribonucleotides, 5 ng / µl of the PH1182 polypeptide obtained in Example 2, and 10 mM Tris-HCl (pH 7.5) was incubated at 37°C for 30 minutes. The reaction product was electrophoresed on a 20% denaturing acrylamide gel (20% acrylamide, 7M urea, 0.5×TBE buffer). After staining with SYBR GREEN II (Takara Bio), the fluorescence image was analyzed with a fluorescence image analyzer FMBIO II Multiview (Takara Bio). The cleavage pattern of each oligoribonucleotide is shown in Table 1.
[0077]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com