Method for recognizing organic estrogen receptor agonism and antagonistic effect
An estrogen receptor and antagonism technology, applied in chemical library, combinatorial chemistry, library screening, etc., to save manpower, material resources and financial resources, low cost and simple effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
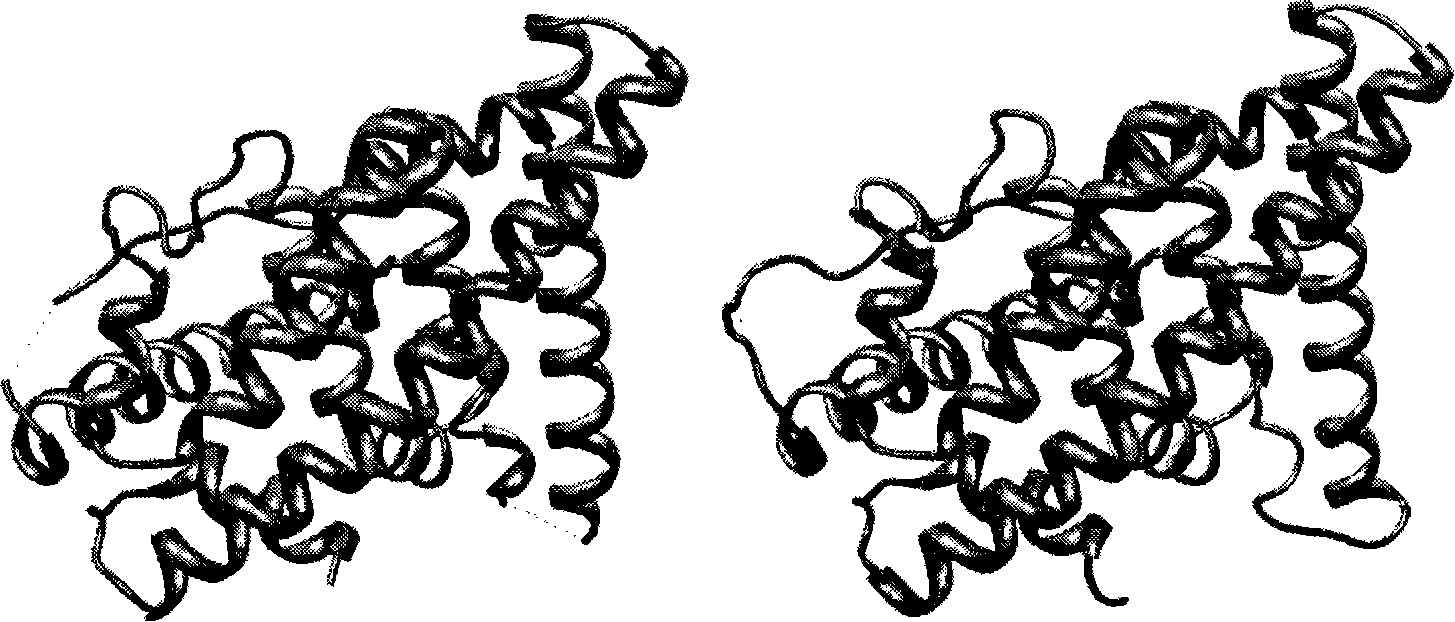
[0031] The method of the invention is used to deal with the problem of complementing the estrogen receptor deletion structure. In the molecular dynamics process, it is necessary to keep the entire protein as a single chain and cannot be composed of multiple peptides. However, in practice, the protein structure obtained in the PDB library often has some amino acid incompletes, which leads to the breakage of the protein chain, and the method of homology modeling is used to fill the missing structure.
[0032] The crystal structure of human estrogen receptor alpha subtype (PDB ID: 1ERE) was used as the initial structure. The structure of 1ERE has deletions in both the loop region between the H1-H2 helices and the loop region between the H9-H10 helices, making the entire 1ERE structure consist of three unconnected peptides. The crystal structure and complete primary sequence of 1ERE were obtained from the PDB library, and the sequence with the crystal structure was compared with th...
Embodiment 2
[0034] Molecular dynamics simulations of complexes are processed using the method of the invention. AMBER cannot automatically identify non-standard amino acids / nucleic acids and ligand molecules. Before performing molecular dynamics simulation on the complex, it is necessary to set the force field of the ligand molecule in the complex. The Antechamber module of AMBER is used to add force field parameters to the ligand molecule. The force field adopts the standard AMBER force field (general AMBER force field, GAFF), and the charge is obtained by the semi-empirical AM1-BCC algorithm. Through Antechamber, the system can identify the atomic bond and atom type information of the ligand, and add the missing force field parameters of the system.
[0035] Use ff03 force field and GAFF force field in tLeap, and load the small molecule force field information into tLeap, then tLeap can read in and correctly identify the complex. Add the necessary Na to balance the charge in the system...
Embodiment 3
[0038] The method of the present invention is used to identify the position of the H12 helix of the typical estrogen-activating ligand 17β-Estradiol (17β-Estradiol).
[0039] The binding mode of estradiol to the ligand-binding domain of the estrogen receptor has been determined experimentally. Select the crystal structure of estrogen and human estrogen receptor α subtype (PDB code 1ERE), and use homology modeling to fill in the missing amino acid residues.
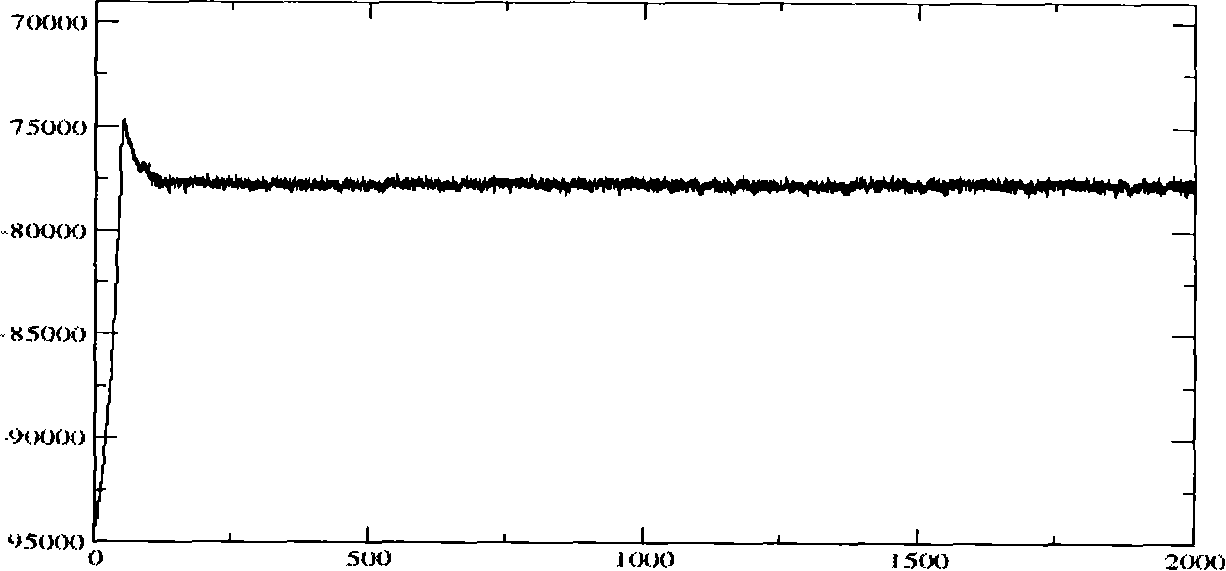
[0040] Molecular dynamics simulations were performed using this structure as an initial model. pass image 3 It can be seen that during the 2ns molecular dynamics simulation, the molecular configuration of the complex has been in a state of equilibrium in a relatively short period of time. During the entire molecular dynamics simulation, the conformation of the complex has not changed much, and the RMSD value has been stable exist . At the same time, it can be seen that after kinetic optimization, the hydrogen bond ne...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com