Medical image diagnosing system and diagnosing method based on migrated nuclear matching tracing
A technology of matching tracking and medical imaging, applied in the field of image processing, can solve the problems of low diagnostic recognition rate and achieve the effect of improving reference value, improving diagnostic recognition rate, and improving generalization ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
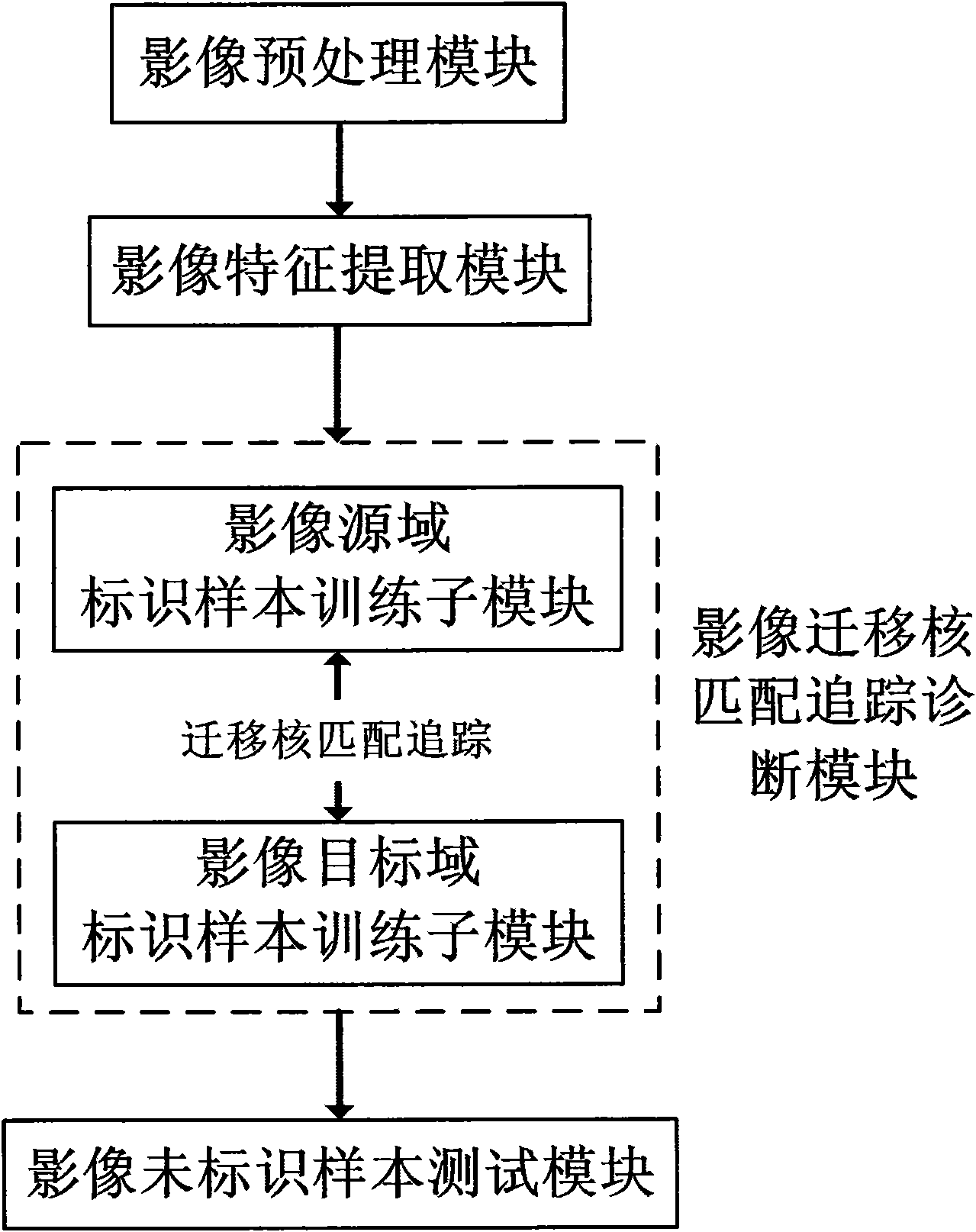
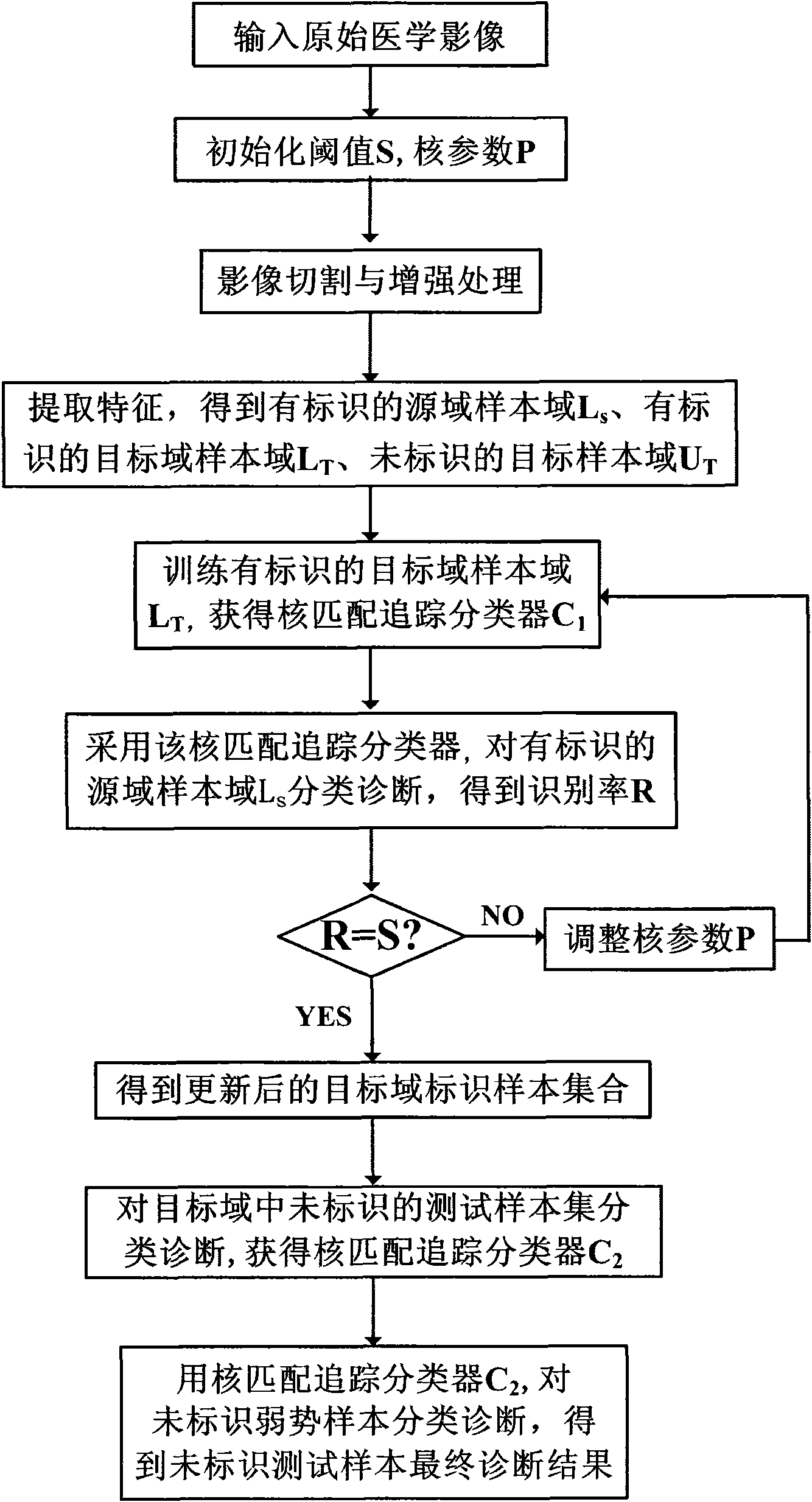
[0035] refer to figure 1 According to the present invention, the medical image diagnosis system based on migration nucleus matching and tracking includes: an image preprocessing module, an image feature extraction module, an image migration nucleus matching and tracking diagnosis module, and an image unlabeled sample testing module. These modules are connected sequentially. The image migration kernel matching tracking diagnosis module includes a training submodule for identifying samples in an image target domain and an identifying sample training submodule for an image source domain. The image target domain identification sample training submodule completes the training of the image target domain with identification samples, generates a training classifier for images with target domain identification samples, and transmits the classifier to the identification sample training submodule of the image source domain; The identification sample training sub-module of the image sour...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com