PCR (polymerase chain reaction) method for detecting Hokovirus for pigs
A hokovirus, downstream technology, applied in the field of animal virology and zoonosis, can solve the problem of disease
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Pig serum and liver collected clinically (Xieqiao Pig Farm, Changshu City, Suzhou City, Jiangsu Province) were tested for pig Hokovirus. The specific steps are as follows:
[0029] (1) Extraction of DNA: Take 472.5 μl of serum; take 100 mg of liver, add 1 ml of PBS buffer for thorough grinding, freeze and thaw three times at -20°C, centrifuge at 8000 rpm for 10 minutes, and take 472.5 μl of supernatant, Add 25 μl of 10% SDS, 2.5 μl of 20 mg / ml proteinase K, mix well and place it at 37°C for overnight digestion or 50°C water bath for 4 hours, add an equal amount of phenol:chloroform:isoamyl alcohol (25:24:1) to pump Extract twice, take the supernatant, add 2 times the volume of pre-cooled absolute ethanol at -20°C for 1 hour, and centrifuge at 12,000 rpm for 10 minutes. The supernatant was discarded, and the precipitate was dried in a drying oven and dissolved in 20 μl of ultrapure water, which was the DNA template for detection. The extracted DNA templates were stored at...
Embodiment 2
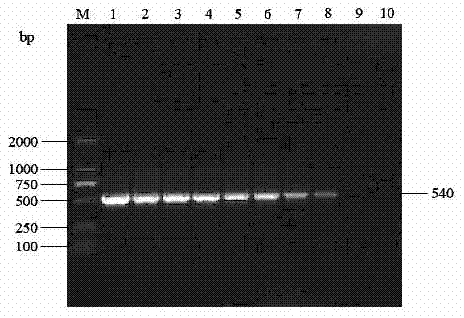
[0034] According to the method of Example 1, the porcine Hokovirus positive disease material is used as a positive control, respectively for porcine reproductive and respiratory syndrome virus, porcine circovirus type 2, swine fever virus, pseudorabies virus, porcine boca virus, porcine transfusion transmitted virus , Japanese encephalitis virus, and porcine Cnvirus were detected, but no bands were amplified, and porcine Hokovirus positive samples amplified a band of about 540 bp in size, see figure 1 . 其序列为SEQ ID NO. 3:GTTGGTCCTGGTAATCCTTTGGATAATGCTCCCGCTAAGGGACCAGTGGATGAAGCAGCGAAACACCACGATGAACGGTACGATGAAATGCTTCGCCATGGTGATTTGCCATACATCCATGGGAGAGGGGCTGATAGGTTGATGAATAAGGAGATAGAGAGGGCGGAGCAGGAGGGTAAAATTGATAACCCTGTGGATGCATTGGTGGGTAATGCAATCCGGGGTATCTGGGAGGCAAAGGAGACTCTTGGGGATATTGCAGATGTTCAATTATCACAGGTTTTACCGCCCGACCCGCCTACCAGCCAAGTGCTTCCGGGCTCTTCAGAAGACGCCCCCAGCCCGAAGAGACAGAGGGCTGGTACCCCTGATTCTGTTCCTACGCCTGGCAACCCATCCCCTGCGCCAATTGCTGATCCTGCTACAATCATGGCTGCGCCGGTAACGGGCGCCACCGGTGGGGGTA...
Embodiment 3
[0036] Example 3: sensitivity test
[0037] The standard positive plasmid containing VP1 / 2 gene (10 11 copies) Use the configured TE (PH8.0) 10-fold incremental dilution as a template, take 2 μl of each dilution as a template, and perform detection according to the method in Example 1, observe positive bands, and use the template used for positive expected bands The highest dilution of the amount was used to calculate its sensitivity, and the results showed that the minimum detection amount was 10 4 copies, see figure 2 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com