Preparation method and application of DNA capturing probe
A technology of capturing probes and magnetic beads, applied in the field of DNA capture probe preparation, can solve the problems of rising probe synthesis cost and high cost of artificial base synthesis method, and achieve the effect of reducing preparation cost and avoiding cost increase.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
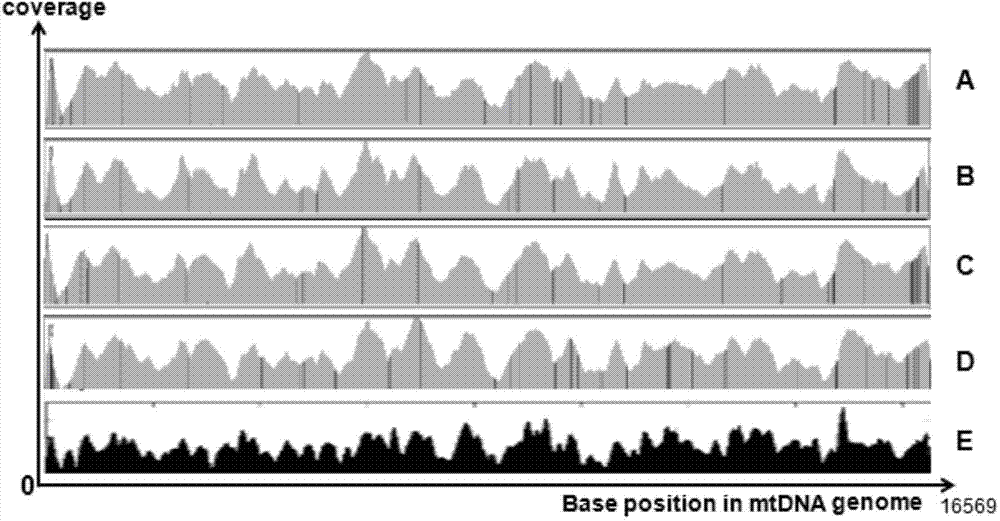
Image
Examples
Embodiment Construction
[0028] The present invention will be described in detail below in conjunction with the accompanying drawings and embodiments.
[0029] (1) The preparation method of human mitochondrial DNA capture probe comprises the following steps:
[0030] 1. Amplify the full-length mitochondrial DNA sequence with three pairs of primers (1F and 1R, 2F and 2R, 3F and 3R);
[0031] The conditions for the amplification are:
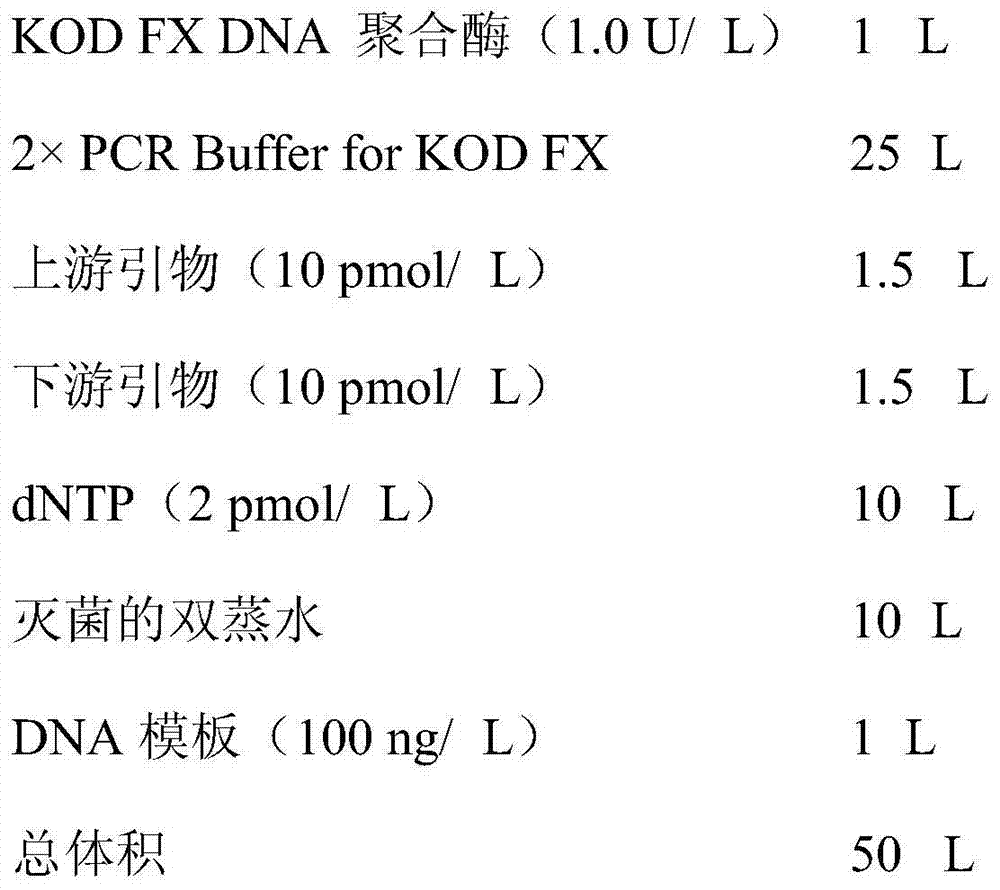
[0032] (1) PCR reaction system:
[0033] The DNA polymerase used is Toyobo KOD FX, product number KFX-101.
[0034] reaction system:
[0035]
[0036] (2) The PCR reaction program was: pre-denaturation at 94°C for 2 min; denaturation at 94°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 8 min; 40 cycles; extension at 72°C for 15 min.
[0037] The sequences of the primers are shown in Table 1:
[0038] Table 1
[0039]
[0040] 2. Mix the amplification products of the three pairs of primers equimolarly, and break them into fragments of about 200bp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com