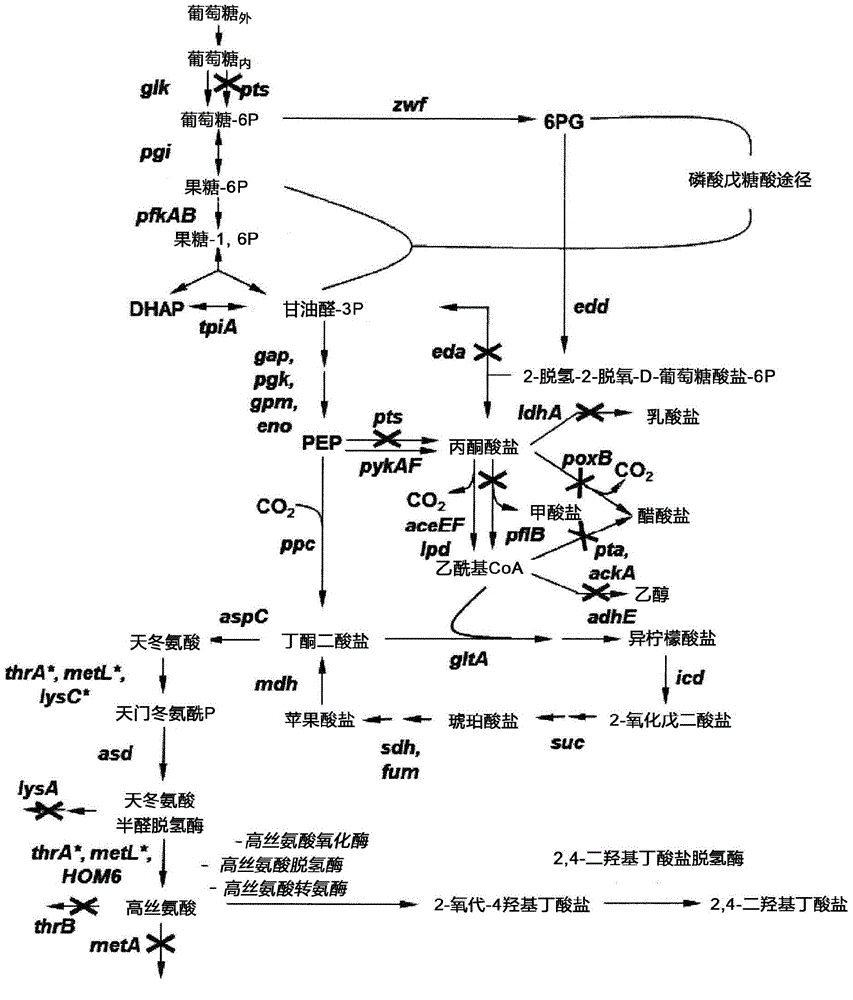
Method for the preparation of 2,4-dihydroxybutyrate
A technology of hydroxybutyrate and carbonyl, which is applied in the field of preparation of 2,4-dihydroxybutyrate, and can solve problems such as undetermined enzyme reactions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1: Demonstration of OHB reductase activity
[0077] Construction of plasmids containing wild-type genes encoding lactate dehydrogenase or malate dehydrogenase:
[0078] Will encode (L)-malate dehydrogenase in Escherichia coli, Ec-mdh (SEQ ID No.1), (D)-lactate dehydrogenase in Escherichia coli, Ec-ldhA (SEQ ID No.3), lactic acid Streptococcus (L)-lactate dehydrogenase, Ll-ldhA (SEQ ID No.5), Bacillus subtilis (L)-lactate dehydrogenase, Bs-ldh (SEQ ID No.7), thermophilic fat The (L)-lactate dehydrogenase of Bacillus, Gs-ldh (SEQ ID No.9), two hypotypes of (L)-lactose dehydrogenase of rabbit, Oc-ldhA (SEQ ID No.11 and The gene of SEQ ID No.13) was extended by PCR using the high-fidelity polymerase Phusion TM (Fermantas) and the primers listed in Table 1, the genomes of Escherichia coli MG1655, Lactococcus IL1403 and Bacillus subtilis chain 168 were used as templates. The genes Oc-ldhA and Gs-ldh were codon optimized for expression in E. coli and synthesized by ...
Embodiment 2
[0093] Example 2: Construction of lactose dehydrogenase with improved OHB reductase activity
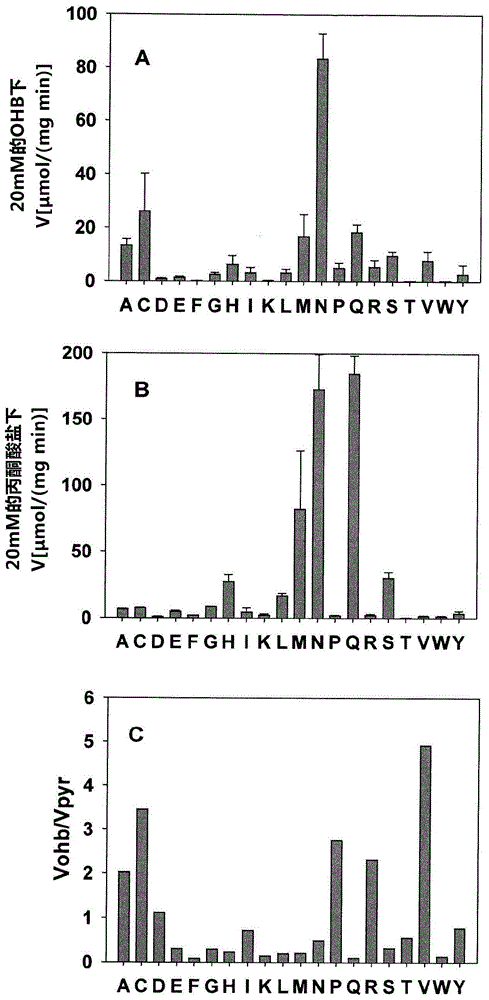
[0094] Site-directed mutagenesis of the lactis ldhA gene was performed using the pET28-Ll-ldhA plasmid as a template. PCR using the oligonucleotide pairs listed in Table 3 (Phusion 1U, HF buffer 20% (v / v), dNTPs 0.2mM, forward and reverse primers 0.04uM, template plasmid 30-50ng, water) to introduce site-directed mutagenesis to alter the amino acid sequence. In addition to functional mutations that facilitate identification of mutant clones, the genes mutated by PCR contained new restriction sites listed in Table 3 (introduced using silent mutations). The template DNA was removed by digesting the PCR product with Dpnl at 37° C. for 1 h, and the PCR product was transformed into competent E. coli DH5α (NEB) cells. Mutated plasmids were identified by restriction site analysis and confirmed to carry the desired mutation by DNA sequencing.
[0095] table 3 : an oligonucleotide used t...
Embodiment 3
[0101] Example 3: Construction of malate dehydrogenase with improved OHB reductase activity
[0102] Site-directed mutagenesis of the mdh gene from E. coli was performed as described in Example 2 using the primers listed in Table 5. Plasmid pET28-Ec-mdh was used as template.
[0103] table 5 Oligonucleotides used to mutate malate dehydrogenase mdh from Escherichia coli (nnk denotes a degenerate codon where k denotes thymine or cytosine)
[0104]
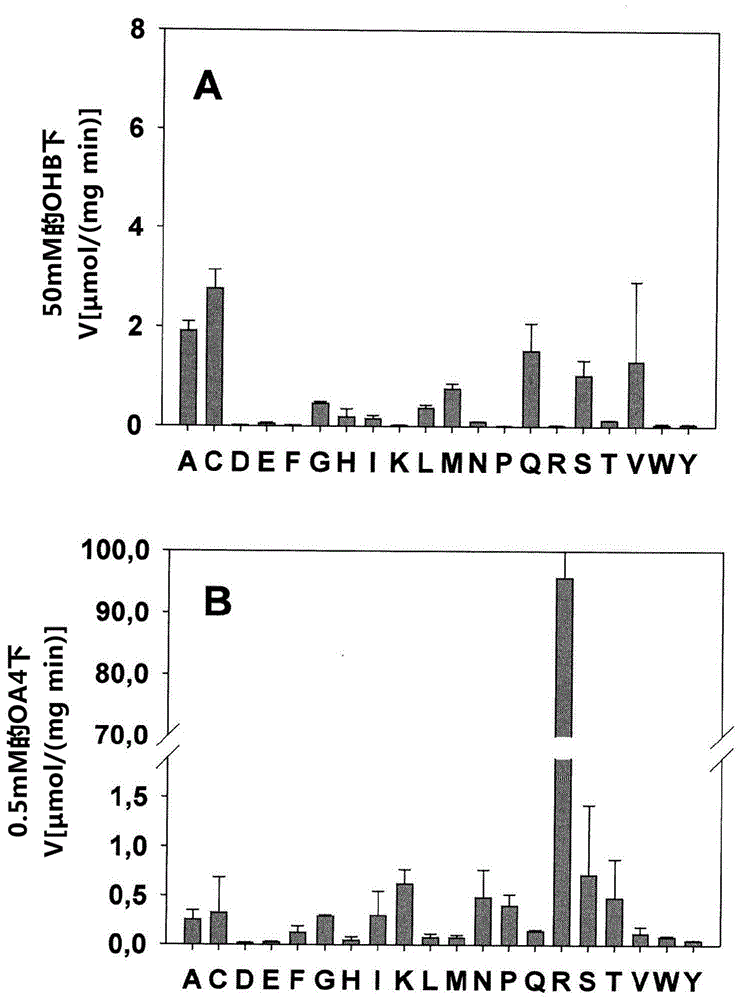
[0105] The mutated enzyme was expressed, purified and tested for OHB and pyruvate reductase activity as described in Example 1. exist image 3 Activity measurements for OHB and pyruvate are summarized in . The results showed that replacement of Arg81 by alanine, cysteine, glycine, histidine, isoleucine, leucine, methionine, asparagine, glutamine, serine, threonine or valine achieved Significant OHB reductase activity, and concomitant decrease in pyruvine reductase activity.
[0106] Combining R81A mutations in Ec-Mdh with o...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap