SNP molecular marker for detecting porcine T cell subset character
A technology of molecular markers and cell subgroups, applied in the direction of recombinant DNA technology, microbial measurement/testing, DNA/RNA fragments, etc., can solve the problem of insufficient abundance of T cell immune-related genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
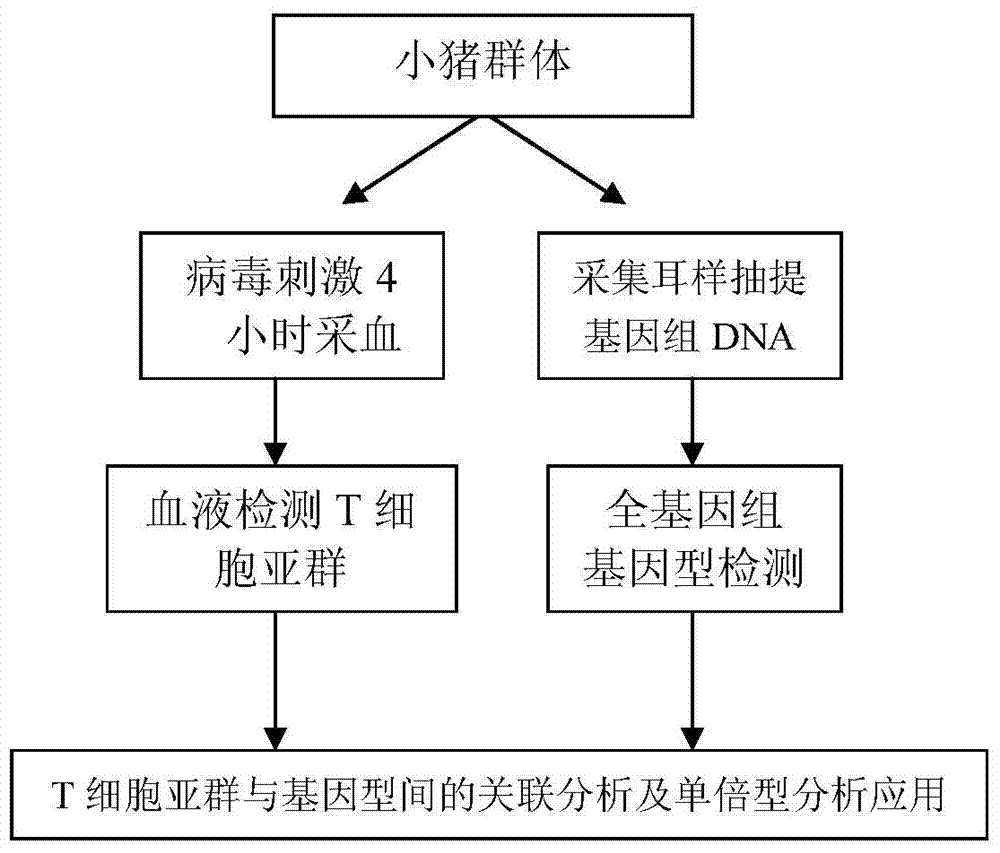
[0032] 1. Sample collection
[0033]The experimental pigs came from 293 piglets of the F2 population obtained by crossing Duroc and Erhualian (obtaining the F2 population obtained by crossing Duroc and Erhualian is a conventional method, the pig breed Duroc is a breed of foreign blood, and Erhualian is a Chinese local Pig blood is a commonly used breed, and the experimental pig breed comes from the experimental pig farm of Huazhong Agricultural University). The pigs have free access to food and water, and the entire feeding method and feeding conditions are always consistent, which is a conventional method.
[0034] Blood was collected from all 35-day-old piglets within 4 hours after Poly I:C injection according to conventional methods.
[0035] 2. Determination of T cell subsets
[0036] The reagents used were mouse anti-pig CD3 monoclonal antibody, mouse anti-pig CD8 monoclonal antibody and mouse anti-pig CD4 monoclonal antibody from Southernbiotech, UK. The main instrume...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com