Molecular marker for detecting wheat fusarium blight QTL Qfsb.hbaas-6BL and application of molecular marker
A technology for Fusarium and wheat, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., and can solve the problem of not developing reliable markers.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1, Discovery of Qfsb.hbaas-6BL and its associated SNP markers
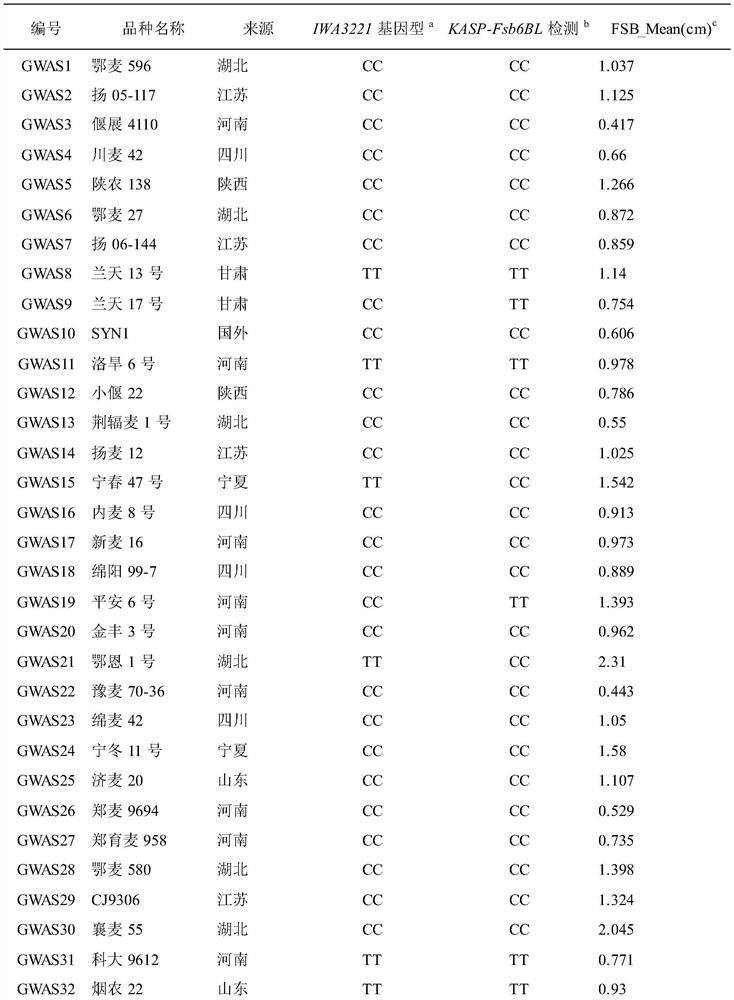
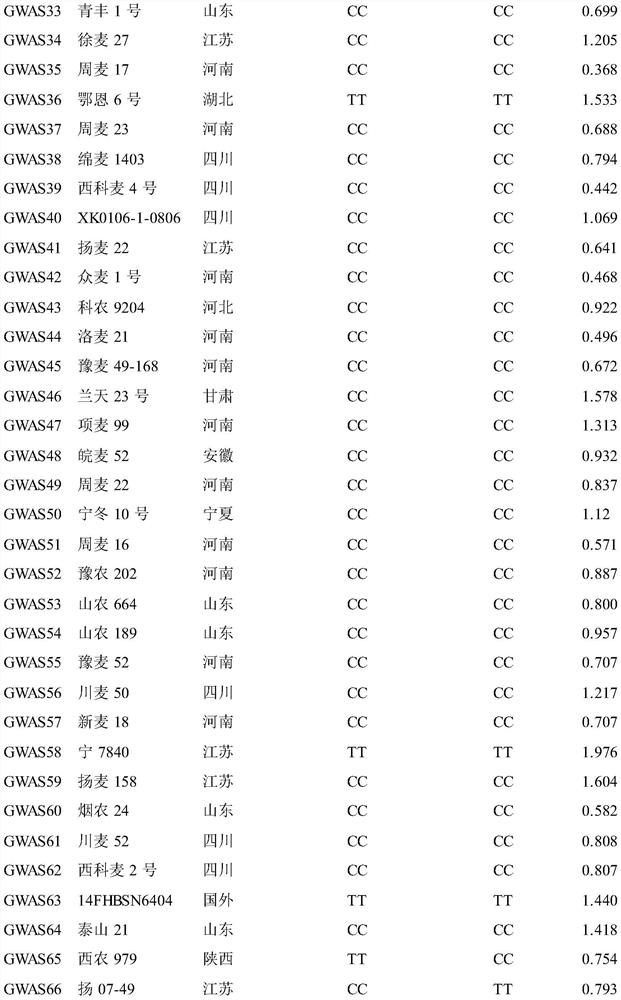
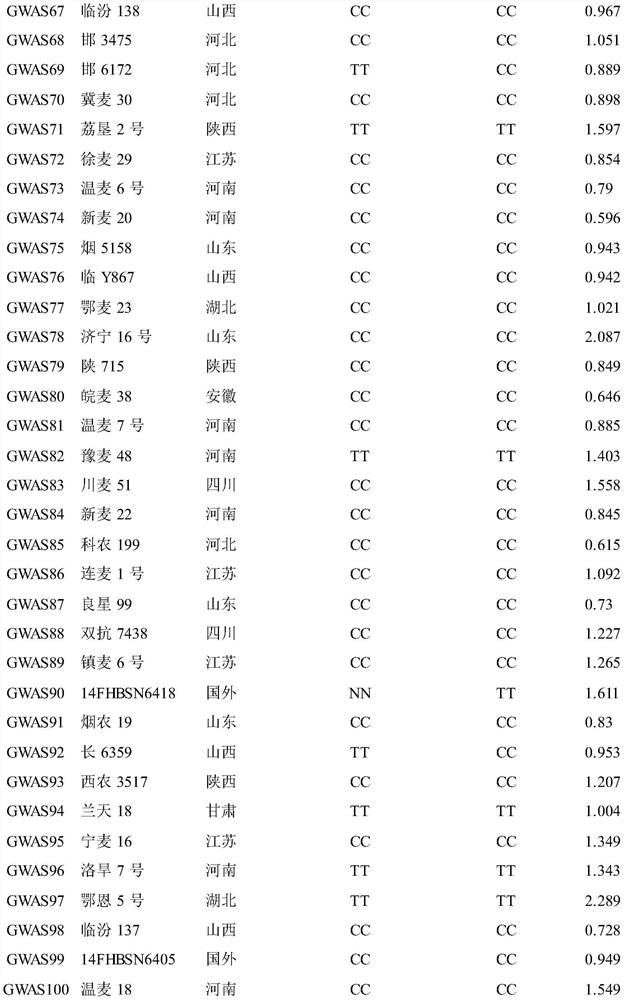
[0045] Test materials: natural populations formed from 240 wheat varieties (lines) at home and abroad, see Table 1. The materials used are recorded in the literature: Zhu Zhanwang, Xu Dengan, Cheng Shunhe, et al. Identification and traceability of the scab resistance gene Fhb1 in Chinese wheat varieties [J]. Acta Crops, 2018, 44(4): 473–482. The public can download from Obtained from the Institute of Food Crops, Hubei Academy of Agricultural Sciences.
[0046]Table 1 240 varieties (lines) and their sources and genotypes
[0047]
[0048]
[0049]
[0050]
[0051]
[0052]
[0053]
[0054]
[0055] a 90K SNP chip typing results; b Mark the detection results for KASP; c Fusarium blight (FSB) lesion length (cm) for 3 repetitions, "NN" means missing chip data.
[0056] 1. Discovery of Qfsb.hbaas-6BL and its associated SNP markers
[0057] 1. Identification of Fusarium bli...
Embodiment 2
[0101] Embodiment 2, actual sample detection
[0102]Test materials: natural population varieties shown in Table 1.
[0103] 1. Identification of Fusarium blight resistance
[0104] With embodiment 1.
[0105] 2. KASP-Fsb6BL marker detection
[0106] Mark with KASP-Fsb6BL according to the method in embodiment 1, carry out KASP reaction to 240 kinds shown in table 1, detect KASP reaction product; Use PHERAstar Plus Detect the product, and use KlusterCaller software for data analysis; if the color is blue, the genotype of the wheat SNP site IWA3221 to be tested is TT; if it is green, the gene of the wheat SNP site IWA3221 to be tested is The type is CC.
[0107] Table 1 shows the marker detection results of 240 wheat cultivars and the average length of FSB lesions. The results in Table 1 were statistically analyzed to obtain the results in Table 5.
[0108] Table 5 Differences in resistance to Fusarium blight of different genotypes of KASP-Fsb6BL
[0109]
[0110] As c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com