Novel application of delta12-fatty acid desaturase gene
A fatty acid dehydrogenase and gene technology, applied in the fields of genetic engineering and biology, can solve problems such as the inability of output and quality to meet market demand, and achieve the effects of low cost, high feasibility and short production cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
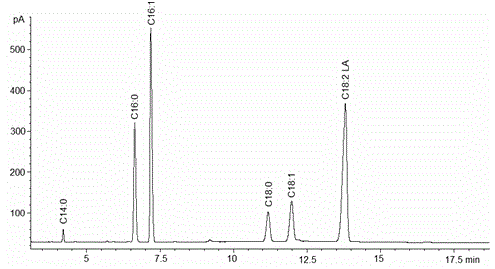
Examples
Embodiment 1
[0015] Example 1: Known and potential Δ for some species of reference yeast 12 - Fatty acid dehydrogenase gene sequence, design specific primers (primer 1 and primer 2), and use cDNA as a template to carry out PCR reaction, the primer sequence is as follows:
[0016] Primer 1: RKD12F:5`-ATAAGCTTATGGCCGCTACCCTCCGCCAGC-3` (SEQ ID NO: 1)
[0017] Primer 2: RKD12R:5`-ATGAATTCCTAGAGTCCCTCGACGCCCGAGT-3` (SEQ ID NO: 2)
[0018] First, use the eukaryotic cell total RNA extraction kit (Promega company product) kit to extract the total RNA of the cell, use the reverse transcription kit (fermentas company product) to reverse transcribe and synthesize the first cDNA, and then use it as a template for PCR Reaction, PCR amplification system (25 μL) is composed as follows:
[0019] 10×Ex Taq Buffer 2.5 μL
[0020] dNTP (2.5 μmol / L) 2 μL
[0021] Template 3 μL
[0022] P1 (10 μmol / L) 2 μL
[0023] R1 (10 μmol / L) 2 μL
[0024] Ex Taq DNA polymerase (5U / μL) 2 μL
[0025] Sterile ddH...
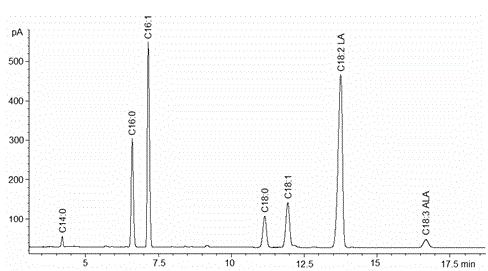
Embodiment 2
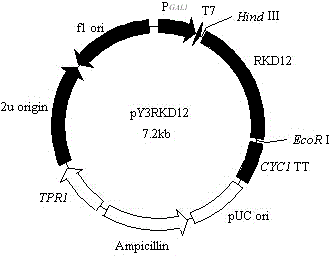
[0027] Embodiment 2: Construction of Saccharomyces cerevisiae recombinant expression vector
[0028] According to the coding region sequence of the sequence obtained in Example 1, a pair of gene-specific amplification primers (primer 3 and primer 4) were designed to isolate its potential open reading frame sequence:
[0029] Primer 3: RKD12F:5`-ATA AGCTT ATGGCCGCTACCCTCCGCCAGC-3`
[0030] Primer 4: RKD12R:5`-ATG AATTC CTAGAGTCCCTCGACGCCCGAGT-3`
[0031] The underlined parts at the 5‵ ends of these two primers respectively contain Eco RI and Hind Ⅲ Restriction site, the amplification conditions and reaction components used are the same as the PCR reaction using cDNA as a template, and the sequencing results of the amplified product show that it is consistent with the sequence of 1-1341bp shown in GenBank accession number KJ502671.1; Then take 25 μl of PCR products and 10ul pYES3 / CT (Invitrogen) for double enzyme digestion, electrophoresis to recover the large fragments,...
Embodiment 3
[0032] Example 3: Saccharomyces cerevisiae recombinant expression vector pY3RKD12 transforms Saccharomyces cerevisiae cells
[0033] Pick a single colony of Saccharomyces cerevisiae strain INVSC1 in 5ml YEPD liquid medium, cultivate it overnight on a shaker at 30°C, transfer it to 100ml of YEPD liquid medium according to the inoculum size of 1%, shake it at 30°C until the OD600 is between 1.3- Between 1.5; 4°C, 4000g centrifugation for 5 minutes to collect the pelleted cells, and use 100ml pre-cooled sterile ddH 2 O Wash the cells, centrifuge at 4°C and 4000g for 5 minutes to collect the pelleted cells, and then use 50ml pre-cooled sterile ddH 2 O Wash the cells, centrifuge at 4°C and 4000g for 5min to collect the precipitated cells, wash the cells with 20ml of pre-cooled 1M sorbitol, centrifuge at 4°C and 4000g for 5min to collect the precipitated cells, suspend the cells with 0.5ml of pre-cooled 1M sorbitol, each 80μl Dispense into 1.5ml pre-cooled centrifuge tubes, take 2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com