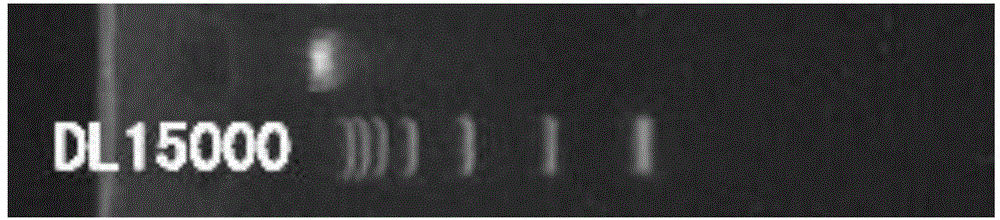
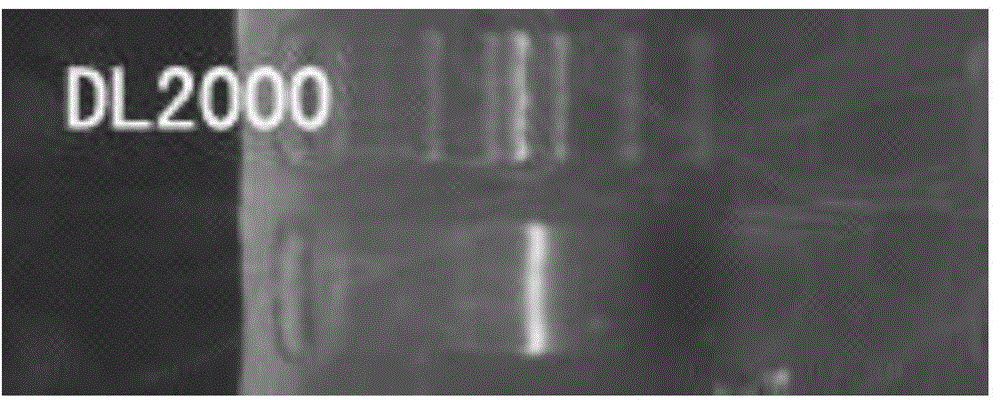
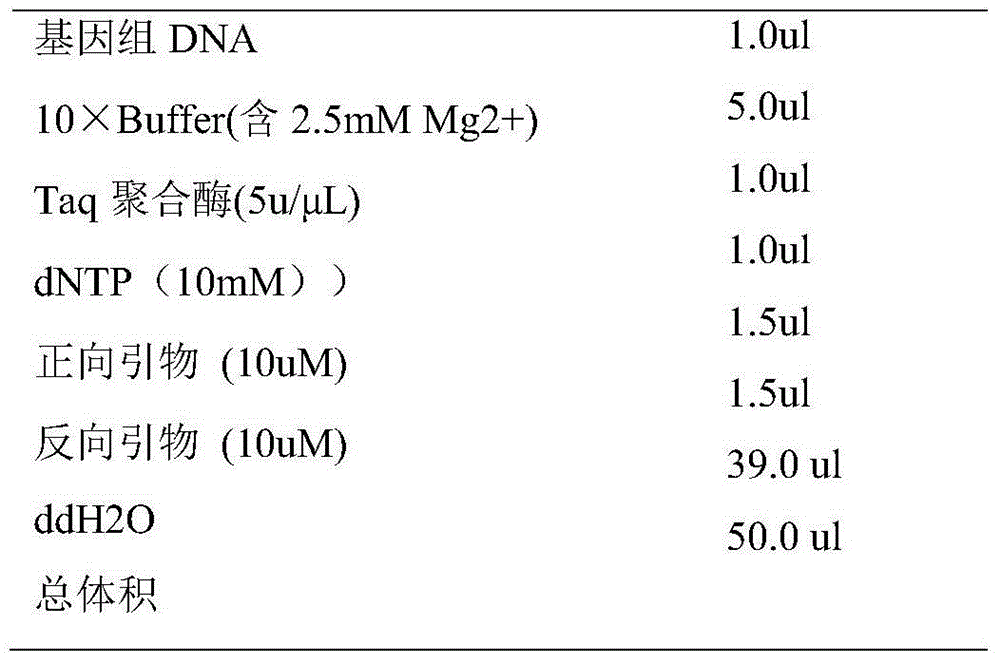
Method for identifying material sources of meat products based on clone and conventional sequencing
A meat product and sequencing technology, applied in the field of molecular biology, can solve the problems of inaccurate and difficult identification results, and achieve the effect of ensuring accuracy and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Material source identification of embodiment 1 canned meat product
[0020] 1. Take the canned meat products, and carry out DNA extraction on the meat products in the cans. The DNA extraction kit of Axygen Biotechnology Co., Ltd. of the United States is used for extraction. The extraction steps are as follows:
[0021] 1) Take 20 mg of meat products, transfer them into a mortar pre-cooled in an ice-water bath, and quickly and vigorously grind them into a homogenate.
[0022] 2) After adding 350 μl Buffer PBS and 0.9 μl RNase A, grind gently for 30 seconds.
[0023] 3) Collect 350 μl of ground tissue homogenate and transfer to a 2 ml centrifuge tube. If the homogenate volume is less than 350 μl, add PBS to 350 μl.
[0024] 4) Add 150 μl Buffer C-L and 20 μl proteinase K. Immediately vortex for 1 min to mix well. After brief centrifugation, place the centrifuge tube in a 56°C water bath for 10 min. (Do not add Proteinase K directly to Buffer C-L).
[0025] 5) Add 35...
Embodiment 2~4
[0052] Examples 2-4 are verified with the identification of known fresh animal tissues, high-pressure processed meat products, and jerky
[0053] Get the fresh animal tissue of known source, high-pressure processed meat product and meat jerky respectively as the meat product sample to be tested, carry out experiment according to the same method steps in Example 1 respectively, verify the accuracy of the method in Example 1 (step are the same, so they will not be repeated), and the COI sequence information of the three meat product samples are respectively obtained as follows:
[0054] Fresh animal tissue:
[0055] The COI sequence (SEQ ID NO:4) is as follows
[0056] AACTGGAGTAAAAGTCTTCAGCTGACTGGCCACTCTGCACGGCGGTGCCATTAAATGAGAAACCCCTCTCCTGTGGGCGCTAGGCTTCATTTTCCTCTTTACAGTTGGAGGATTGACCGGAATCGTTCTAGCTAATTCTTCTCTAGACATTATGCTTCACGACACATATTATGTCGTCGCCCACTTCCACTATGTCCTCTCAATAGGAGCCGTTTTCGCCATTGTTGCCGGCTTCGTCCACTGATTCCCCCTATTCTCAGGGTACACGCTTCACGACACTTGAACAAAAATCCACTTTGGGGTTATATTTGTAG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com