A method for fixing targeting primers in single-molecule targeted sequencing, a single-molecule targeted sequencing kit, and applications
A fixed method and targeted sequencing technology, applied in the field of molecular biology, can solve the problem that the fixed density is only 100-200/field of view.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment
[0059] Material and Reagent Description:
[0060] Unless otherwise specified, the reagents used in the examples of the present invention are commercially available, and the databases used in the examples of the present invention are all public online databases.
[0061] 20x SSC: Dissolve 175.3g NaCl and 88.2g sodium citrate in 800ml water, add a few drops of 10mol / l NaOH solution to adjust the pH to 7.0, add water to 1L, aliquot and autoclave. SSC is the most standard blotting and molecular hybridization treatment solution in molecular biology, and 20xSSC is a common concentrated buffer used for denaturation and cleaning of hybridization experiments.
Embodiment 1
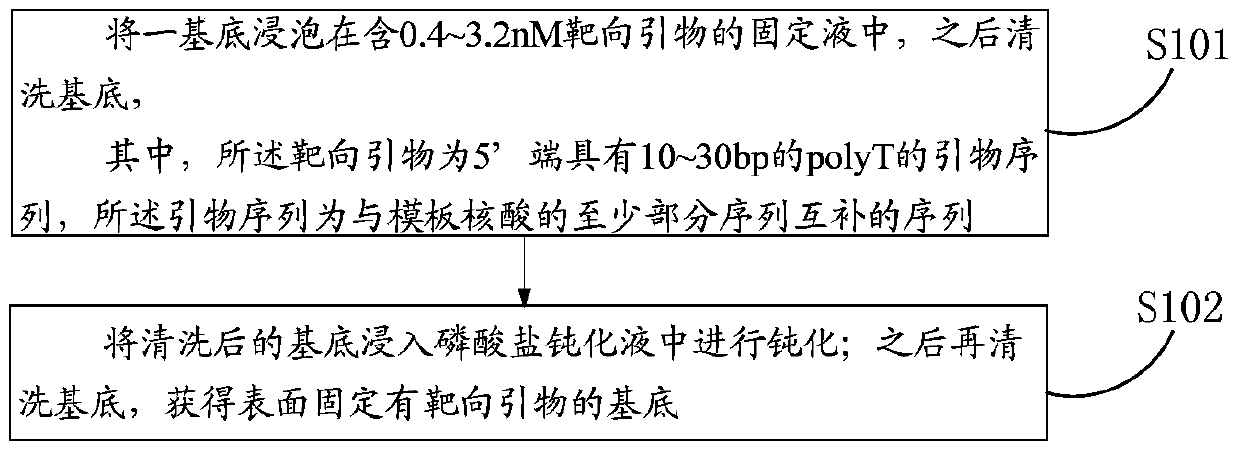
[0063] This embodiment provides a method for fixing targeting primers in single-molecule targeted sequencing, which specifically includes the following steps:
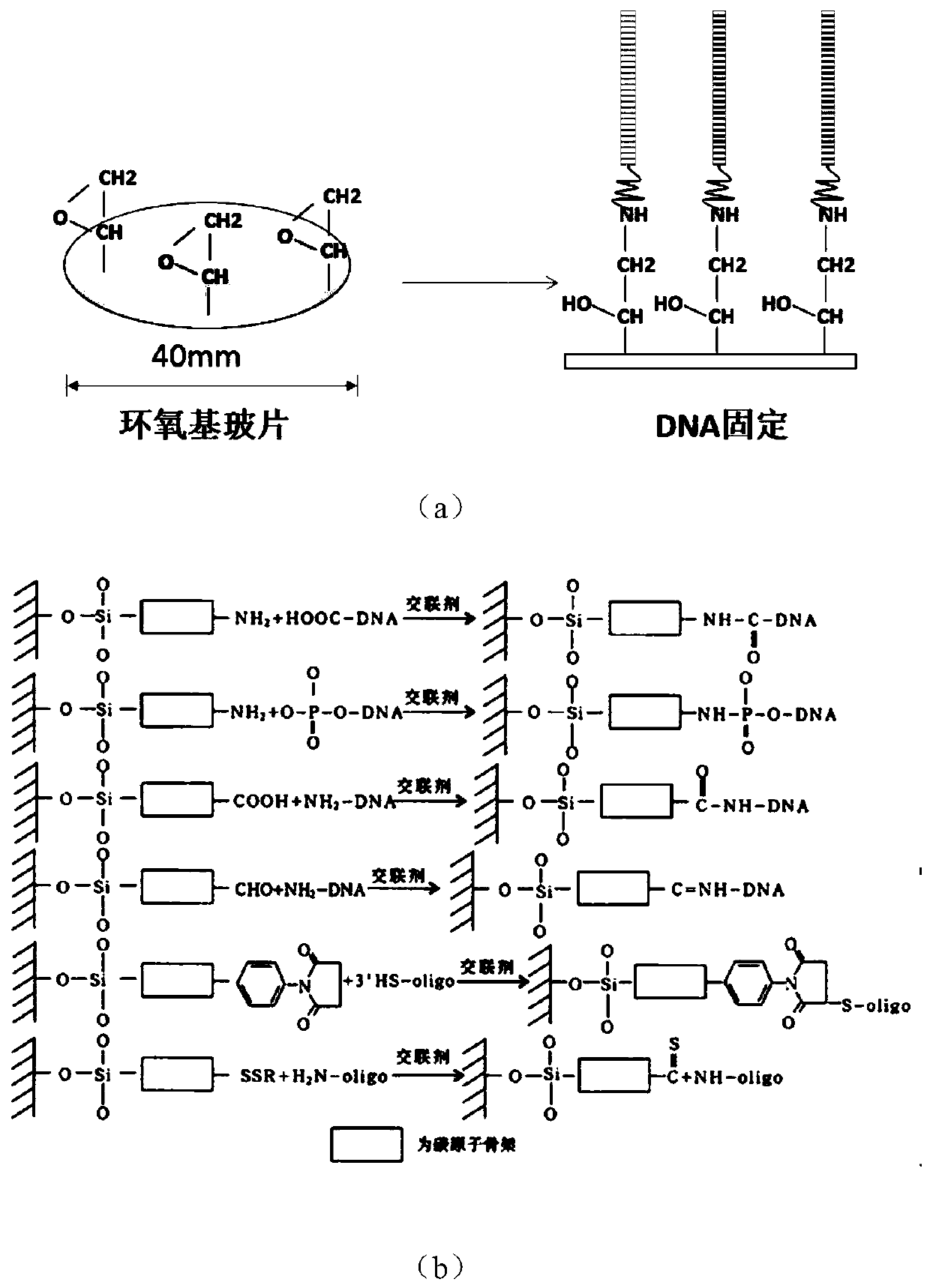
[0064] (1) Blow off the substrate to be fixed with a nitrogen gun for use, the surface of the substrate has an epoxy group, and the substrate slide is from an epoxy-modified slide of SCHOTT;
[0065] With 0.2M of K 2 HPO 4 Dilute the targeting primer to a concentration of 0.8nM in the fixing solution, soak the substrate in the above fixing solution containing 0.8nM targeting primer, soak for 60min;
[0066] Afterwards, the mixed solution composed of 3x SSC and 0.1% Triton, 3x SSC and K with a concentration of 150 mM and pH=8.5 were used successively. 2 HPO 4 to clean the base;
[0067] (2) Immerse the cleaned substrate into a phosphate passivation solution, the phosphate passivation solution is pH=9, and the concentration is 1M K 2 HPO 4 solution, and passivated on a shaker at room temperature at a speed of 80rpm...
Embodiment 2
[0130] The effect of shaking speed in passivation on the fluorescent spots in fixed photos and the effect of different concentrations of targeting primers on the fixation effect
[0131] Operate according to the experimental method described in above-mentioned embodiment 1, difference is:
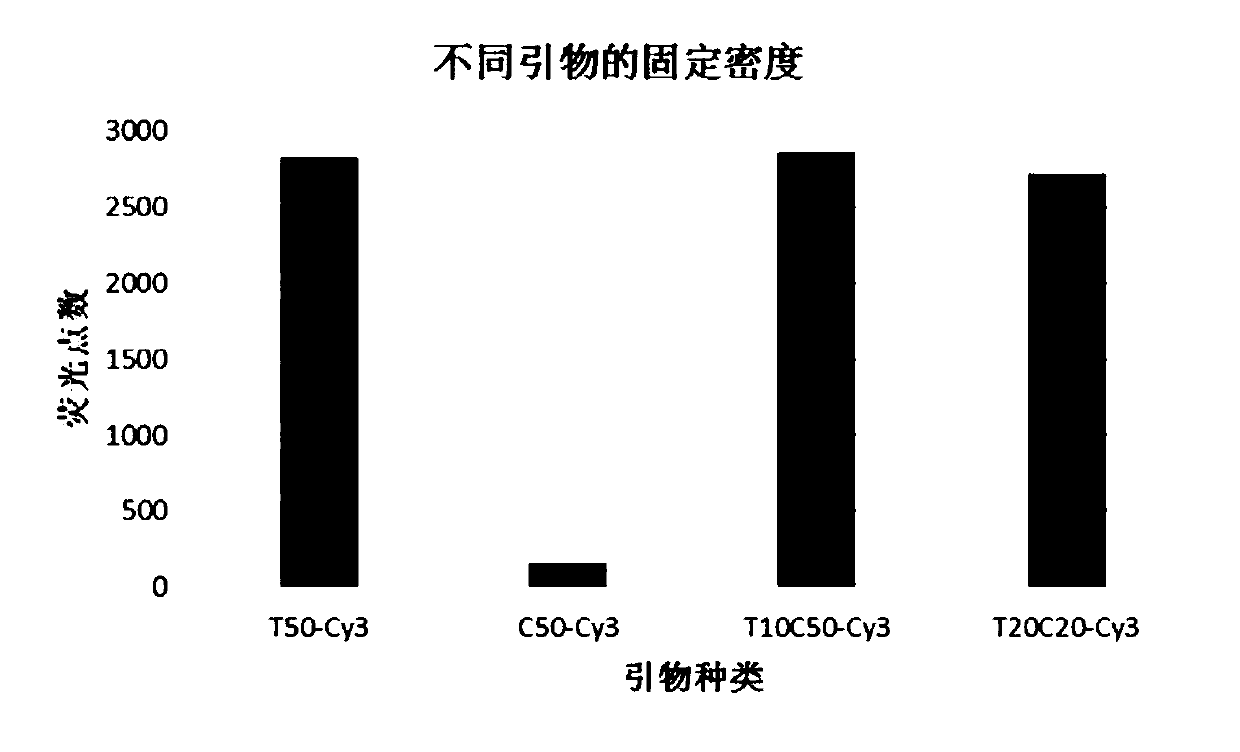
[0132] In step 1), the targeting primer to be immobilized is T10C50-Cy3, and its concentrations are 0.4nM, 0.8nM, 1.6nM, 3.2nM respectively; in addition, in step 2), when passivating, divide into two batches and carry out parallel In the test, two groups of experiments were passivated for 15 hours under shaking conditions of 0rpm / min and 80rpm / min respectively.
[0133] Repeat three times and count the number of sequences immobilized on the substrate. The results of the fluorescence microscope are as follows: Figure 4 and Figure 5 shown. Figure 4 This is the result of fluorescence microscopy photos taken during the passivation process when the rotational speed is 0 rpm / min, including ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com