Methods for genome assembly and haplotype phasing
A genome assembly and haplotype technology, applied in biochemical equipment and methods, computer combinatorial chemistry, determination/inspection of microorganisms, etc., can solve problems such as difficulty in generating high-quality and highly contiguous genome sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0245] Example 1. Methods of producing chromatin in vitro
[0246] Two approaches to remodel chromatin deserve special attention: one approach uses ATP-independent random deposition of histones onto DNA, and the other approach uses ATP-dependent assembly of periodic nucleosomes. The present invention allows any one approach to be used in conjunction with one or more of the methods disclosed herein. Examples of the two ways to generate chromatin can be found in Lusser et al. ("Strategiesforthereconstitution of chromatin", Nature Methods (2004), 1(1): 19-26), the entire contents of which are incorporated herein by reference, including the references cited therein literature.
Embodiment 2
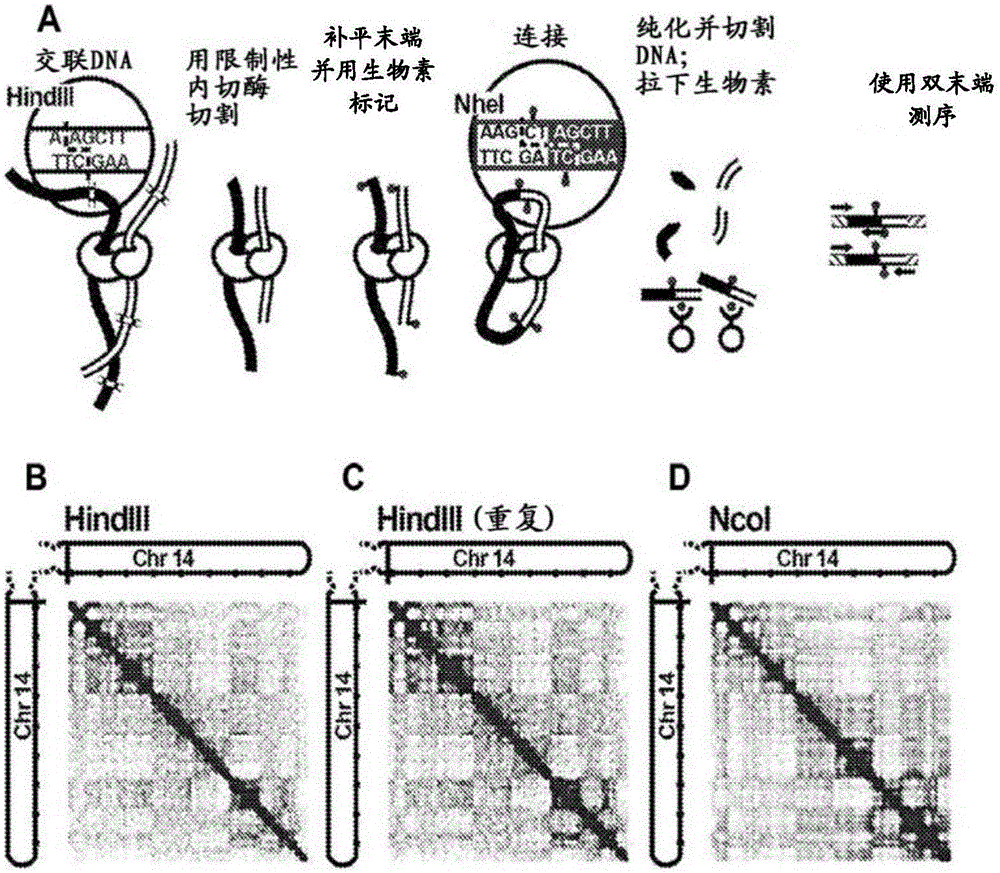
[0247] Example 2. Genome assembly using HI-C-based technology
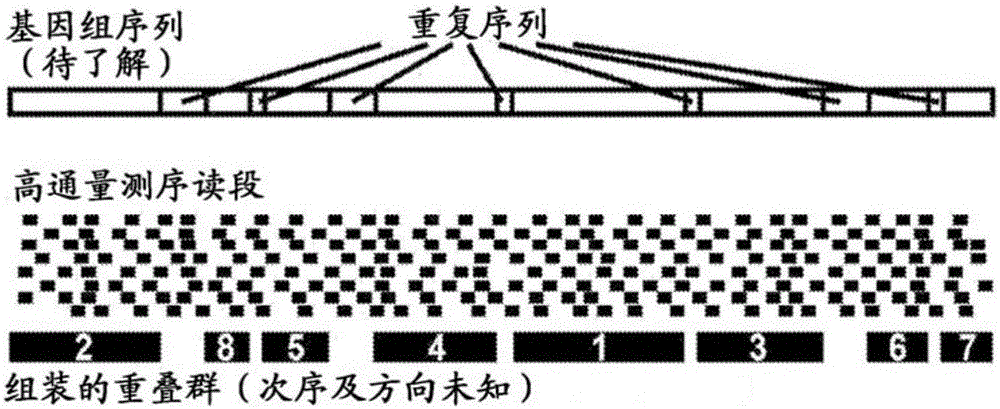
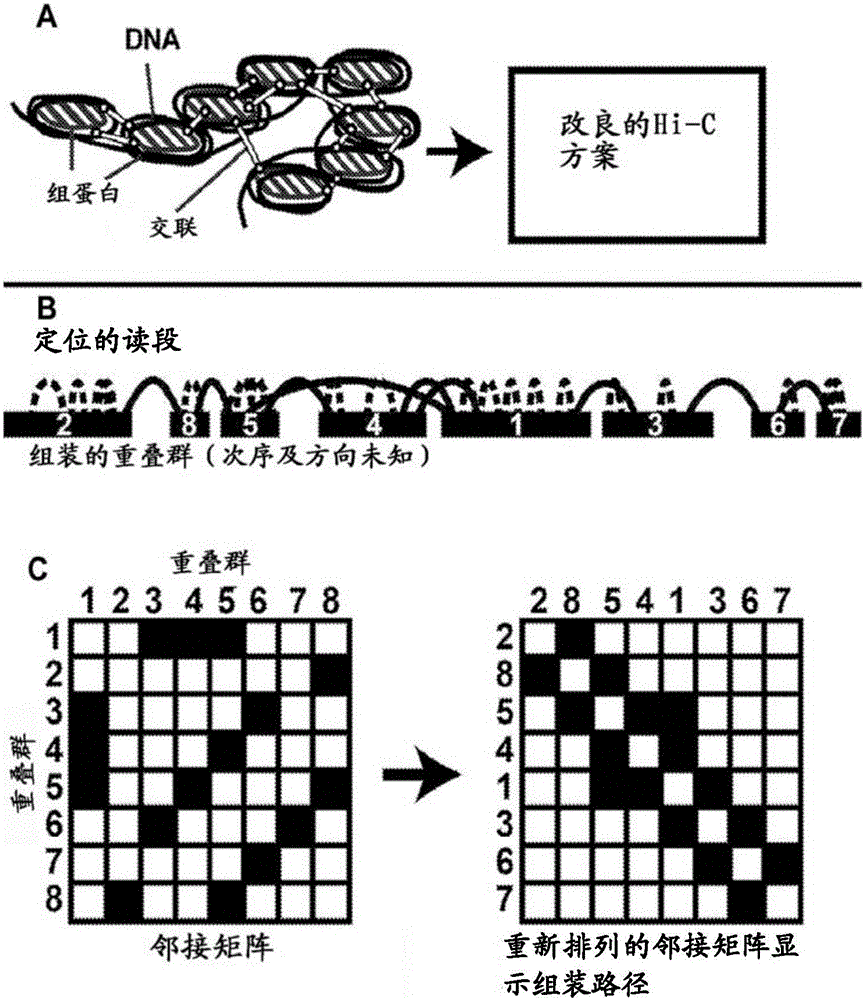
[0248] The genome from a human subject was fragmented into pseudo contigs of 500 kb in size. Using the Hi-C-based method, multiple read pairs are generated by probing the physical layout of chromosomes in living cells. A variety of Hi-C-based methods can be used to generate read pairs, including the method shown below: Lieberman-Aiden et al. ("Comprehensive mapping of longrangeinteractionsrevealsfoldingprinciplesofthehumangenome", Science (2009), 326(5950):289-293), the entire contents of which are quoted The method is incorporated herein, including the references cited therein. Read pairs are located to all pseudo contigs, and those read pairs located to two independent pseudo contigs are used to construct an adjacency matrix based on the positioning data. By using the function of the distance from the reads to the edge of the pseudo contig, the read pairs of about 50%, about 60%, about 70%, about 80%, about 90%,...
Embodiment 3
[0253] Example 3. Method for haplotype phasing
[0254] Because the read pairs generated by the methods disclosed herein are usually derived from intrachromosomal contacts, any read pair containing a heterozygous site will also carry information related to its phasing. Using this information, reliable phasing can be performed quickly and accurately at short, medium, or even long (million base) distances. Designed for an experiment to phase data from one of 1,000 genome triads (a collection of maternal / paternal / offspring genomes) to reliably infer phasing. In addition, haplotype reconstruction can also be used in conjunction with the haplotype phasing method disclosed herein, which uses ortho-linkage similar to Selvaraj et al. (Nature Biotechnology 31: 1111-1118 (2013)).
[0255] For example, haplotype reconstruction based on the ortho-linkage method can also be used in the methods disclosed herein for phasing the genome. Haplotype reconstruction using a method based on proximity...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com