A key marker site affecting the content of c20:0 saturated fatty acid in pig muscle and its application
A site and marker technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve problems such as the inability to determine the key variation sites of the main gene, and the breeding and improvement of difficult breeding pigs, etc. Achieve the effect of optimizing fatty acid composition, high quality and improving nutritional level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Detection of 60,000 (60K) SNP genotypes in the whole pig genome: from the above F 2 A small piece of ear tissue sample was collected from each individual in the resource group and Sutai pig group, and the whole genome DNA was extracted by the standard phenol-chloroform method. After the quality was tested by the Nanodrop-100 spectrophotometer, the concentration was uniformly diluted to 50ng / ul. On the Illumina Beadstation platform, the pig whole genome 60K SNP (Illumina, USA) genotype was determined according to the company's standard process. Use PLINK (1.07) to perform quality control on the 60K chip data of all samples, and eliminate individuals whose detection rate is lower than 95%, and whose family Mendelian error rate is higher than 0.1; and the detection rate is lower than 95%, and the minimum allele frequency is less than 0.05 and Hardy-Weinberg equilibrium significance above 10 -6 SNP markers.
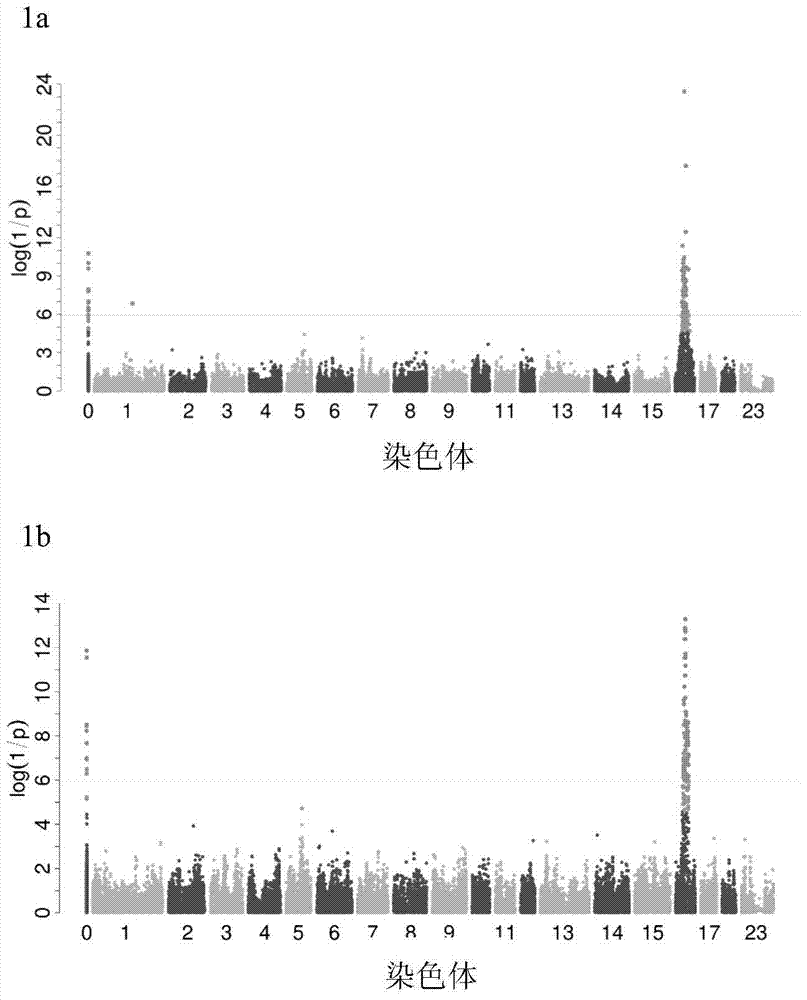
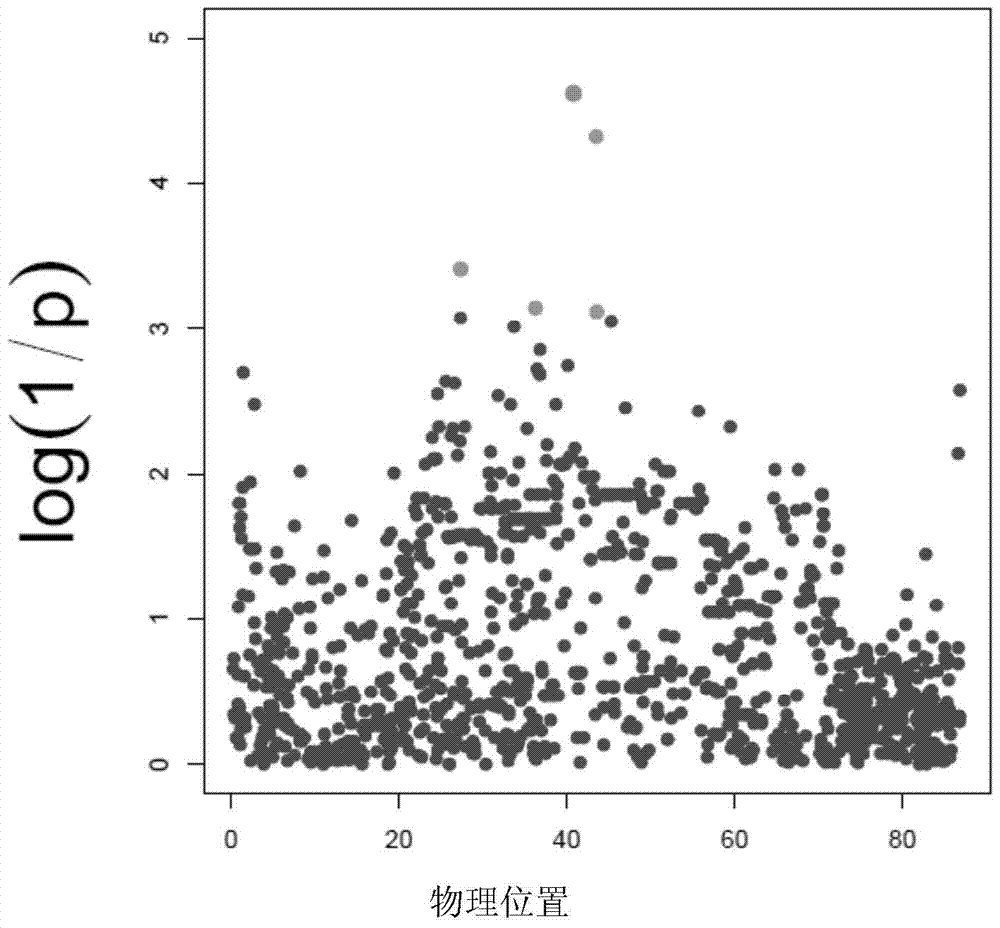
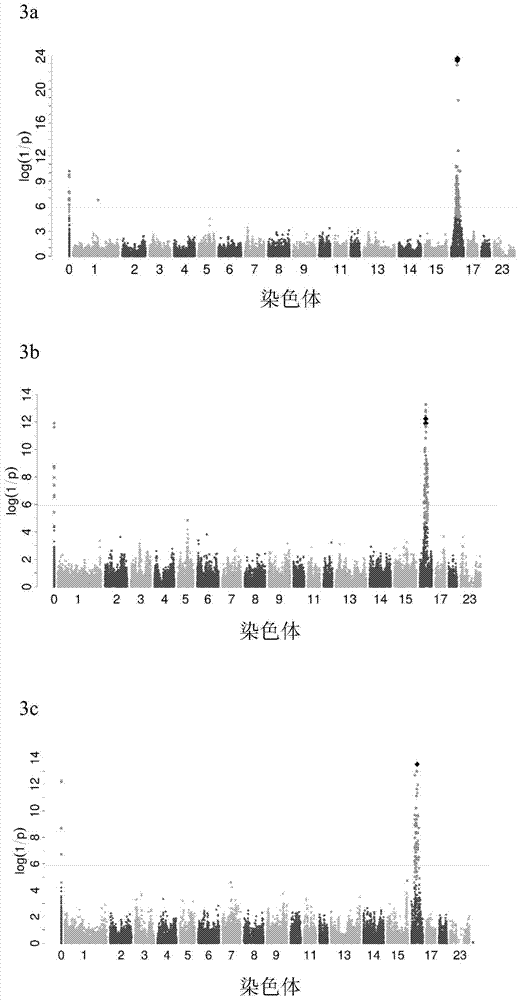
[0045] Genome-wide association (GWAS) analysis, fine mapping an...
Embodiment 2
[0049] This example is the application of the SNP sites g.42561841G>A and / or g.42564441C>T obtained in Example 1 in breeding pigs. SNP locus g.42561841G>A typing: Using the genomic DNA of F2, Sutai population, Laiwu population, Erhualian population and Du Dachang business population as templates, ABI 7900 real-time fluorescent quantitative PCR system was used for genotyping. The reaction system is as follows: DNA template 1 μl, front primer (GGATGAATGGCATAGATAAAAAGATG) 0.2 μl, back primer (GGTAGATCGCAATCCTTCAGTTTC) 0.2 μl, SEQ ID NO: 2 0.15 μl, SEQ ID NO: 30.15 μl, TaqProbe Mix 5 μl, add ddH2O to make up to 10 μl; The conditions are as follows: pre-denaturation at 50°C for 2min; denaturation at 94°C for 10min, 95°C for 15s, 60°C for 15s, 40 cycles. Only VIC fluorescence indicates that the allele type is homozygous GG; only FAM fluorescence indicates that the allele type is homozygous AA; both VIC and FAM fluorescence indicate that the allele type is heterozygote GA.
[0050] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com