CMV neutralizing antigen binding proteins
A technology that binds proteins and antigens, and is applied in antiviral immunoglobulins, immunoglobulins, antibodies, etc., and can solve problems such as CMV infection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0293] Example 1: Production of a set of rabbit monoclonal antibodies
[0294] The expression of the pentameric gH complex in AD169 was restored and the revertant virus was able to infect MRC-5 and ARPE-19 cells. AD169 (GenBank Accession No. X17403) from ATCC is propagated in MRC-5 cells (Fu et al., 2012, Vaccine 30: 7469-7474; Tang et al., 2011, Vaccine 29: 8350-8356). As described in Fu et al. (2012, Vaccine 30: 7469-7474) and Tang et al. (2011, Vaccine 29: 8350-8356), the revertive virus was generated by continuous passage adaptation of AD169 in culture. The AD169 virus and its revertant isolate were amplified in MRC-5 (ATCC accession number CRL-171) or ARPE-19 (ATCC accession number CRL-2302).
[0295] As determined by antigen titration enzyme-linked immunoassay (EIA), both the revertant virus and its parent AD169 virus contained the same level of gB. Figure 1A Show gB monoclonal antibody B8.6 (IgG 2a κ) and 35.1 (IgG 2a κ) (both developed endogenously) is equivalent to the ...
Embodiment 2
[0298] Example 2: Binding and neutralization profile of anti-CMV monoclonal antibodies
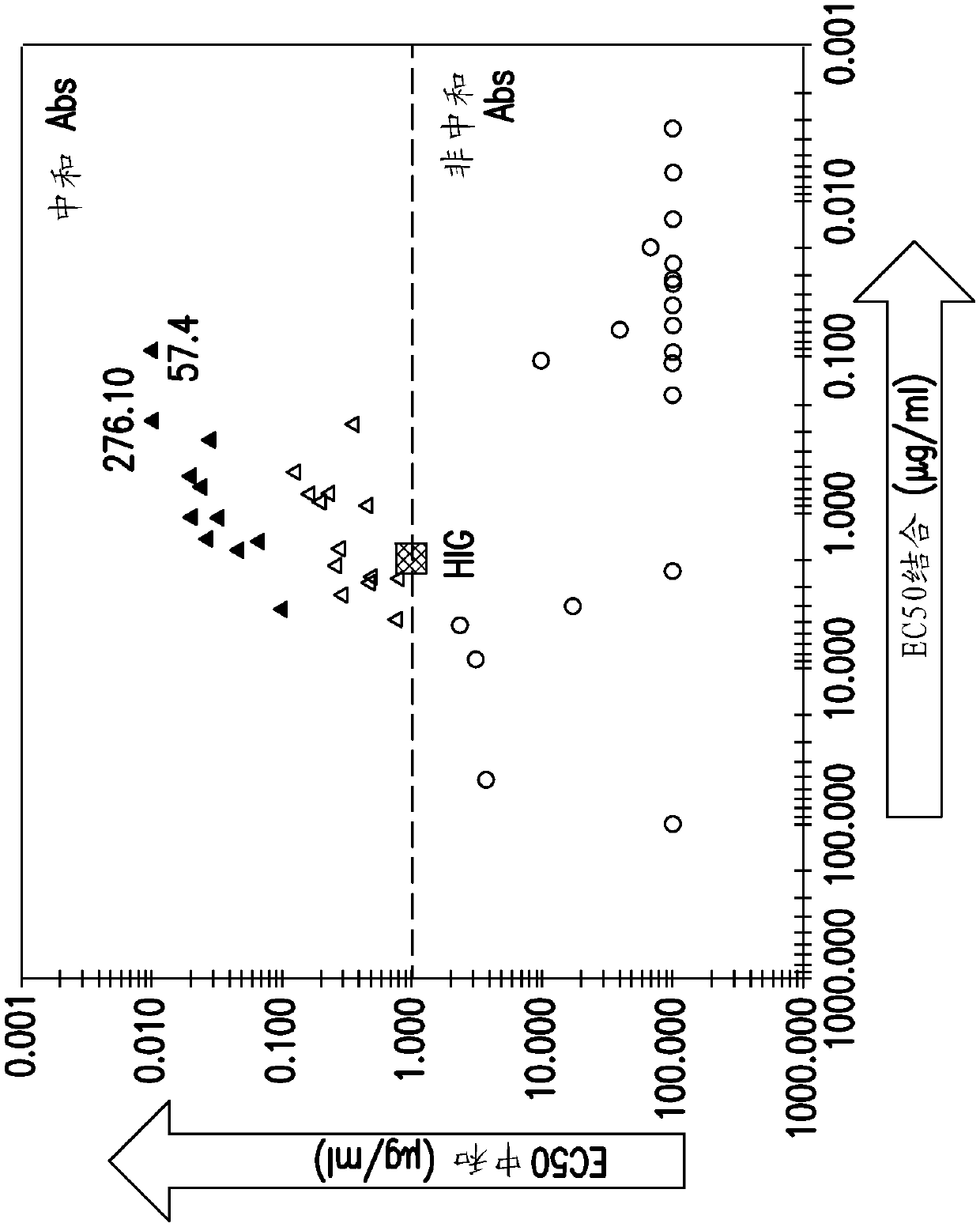
[0299] The ability of the monoclonal antibody to neutralize and bind to the virus and neutralize was determined. The neutralization assay assesses the ability of monoclonal antibodies to prevent entry of viral epithelial cells. The virus binding assay uses ELISA to assess the ability of monoclonal antibodies to bind to virions.
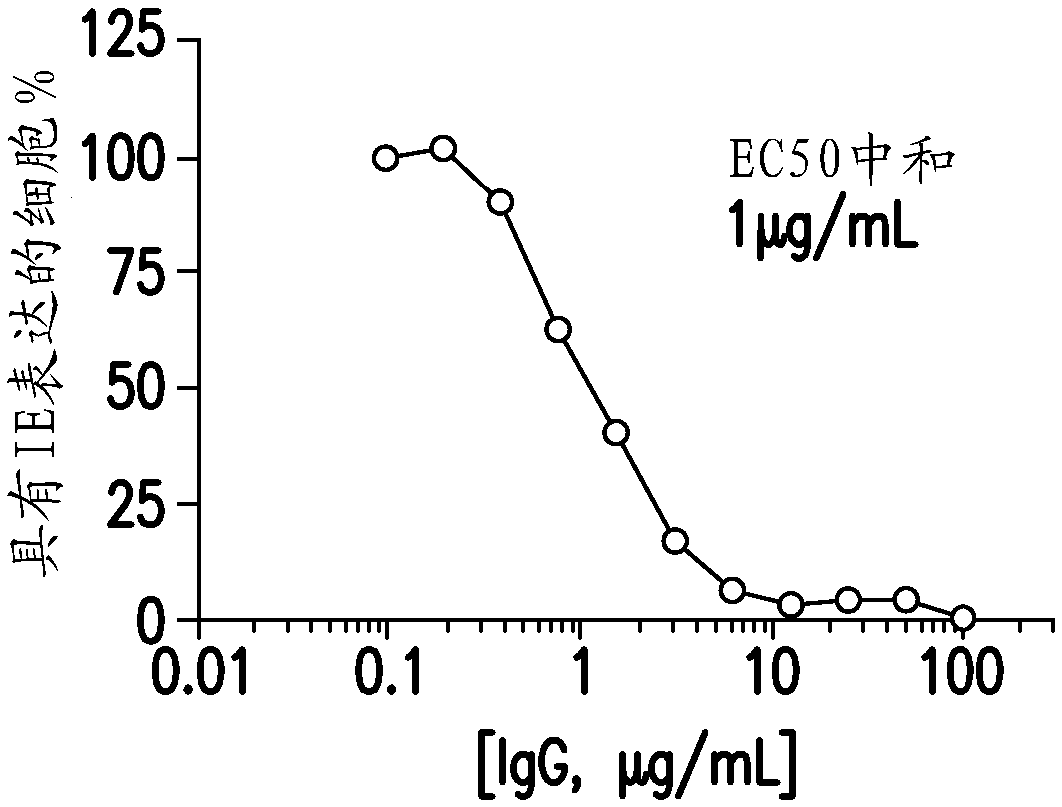
[0300] Briefly, the neutralization assay used is based on counting cells expressing viral immediate early (IE) antigen 24 hours after infection and as previously described (Tang et al., 2011, Vaccine 29: 8350-6). Use Prism 5 (GraphPad Software, SanDiego, CA) to calculate EC from four-parameter curve fitting 50 Value, which is defined as the concentration of antibody required to block 50% of the virus entry.
[0301] In short, the virus binding assay used is an antibody titer enzyme-linked immunoassay (EIA) to determine the relative binding affinity of each monoclonal ...
Embodiment 3
[0312] Example 3: The neutralizing ability of antibodies in epithelial cells is not related to their activity in fibroblasts
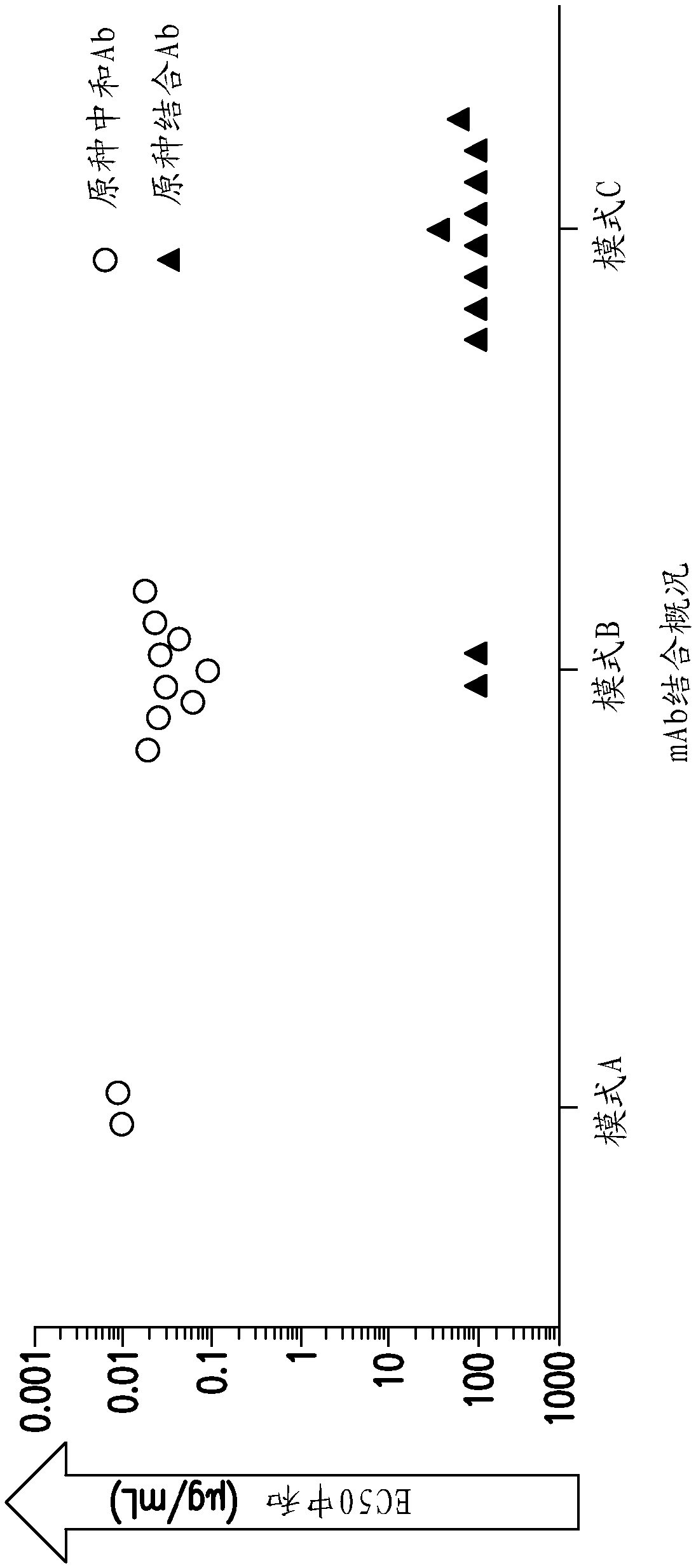
[0313] It is known that HIG can block the entry of viruses into fibroblasts, such as MRC-5 cells, and is about 10 to 15 times less effective than blocking the entry of viruses into epithelial cells, namely ARPE-19 cells (Cui et al., 2008, Vaccine 26: 5760-5766). It has been suggested that a different entry mechanism is used for infecting epithelial cells compared to fibroblast viruses (Wang et al., 2007, PNAS 104: 20037-42). Therefore, by measuring the EC in each monoclonal antibody MRC-5 cell 50 Neutralize to evaluate the antibody group (Table 5). Antibody blocks the virus from entering MRC-5 cells (y-axis) compared to the EC of ARPE-19 cells (x-axis) 50 The relationship between the values is shown in image 3 in. All 45 kinds of antibodies can be classified into three groups: the antibodies in group A only neutralize the virus in ARPE-19 cells, the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com