Molecular marker combination for Litopenaeus vannamei germplasm identification and application thereof
A molecular marker, Vanabine technology, applied in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problem of distinguishing different varieties, difficult to guarantee the authenticity of varieties and seedlings, disrupting the market operation order, etc. problem to improve the accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Development of a method for identification of Litopenaeus vannamei germplasm by combination of microsatellite markers
[0027] 1. Source of materials: Collection of germplasm materials from Zhengda, Kona Bay, SIS, Molokai, Ecuador, Kehai No. 1, and Guihai No. 1. Bay includes materials from 2012, 2014 and 2015, SIS includes materials from 2015 and 2016, materials from Guihai 1, Kehai 1, Molokai and Ecuador from 2014, the total number of samples 192.
[0028] 2. DNA extraction: The DNA of the above-mentioned materials was extracted using the Tiangen Plant Genome Extraction Kit. For the method, refer to the kit instructions. The concentration of the extracted DNA was measured using a Nanodrop 1000 (Thermo, USA), and stored in a -20°C refrigerator.
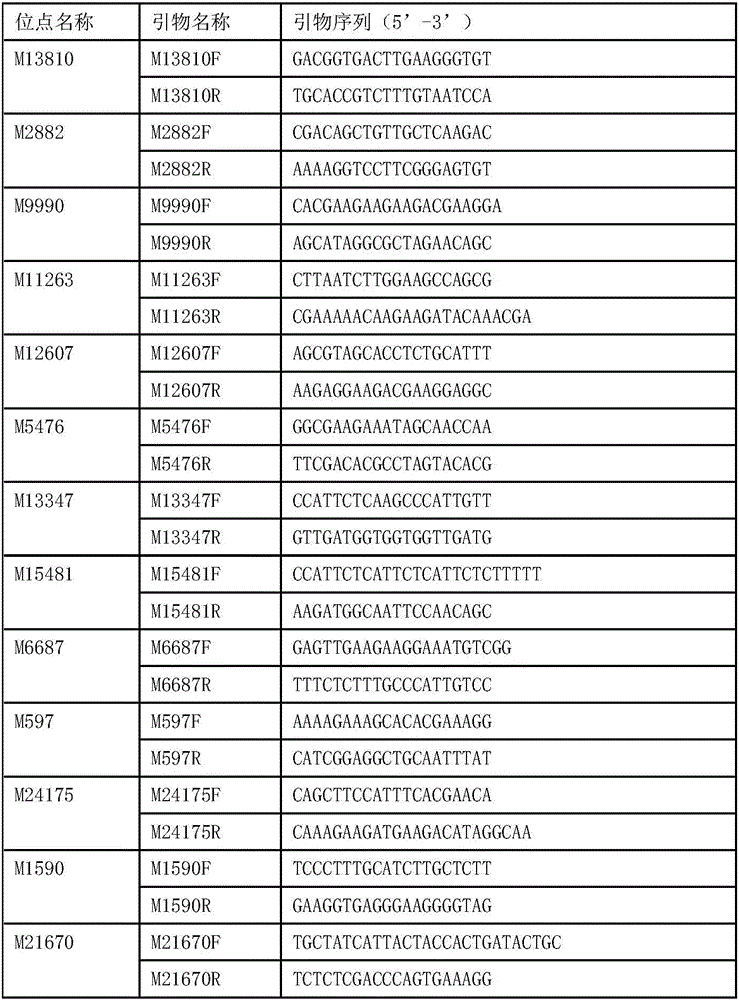
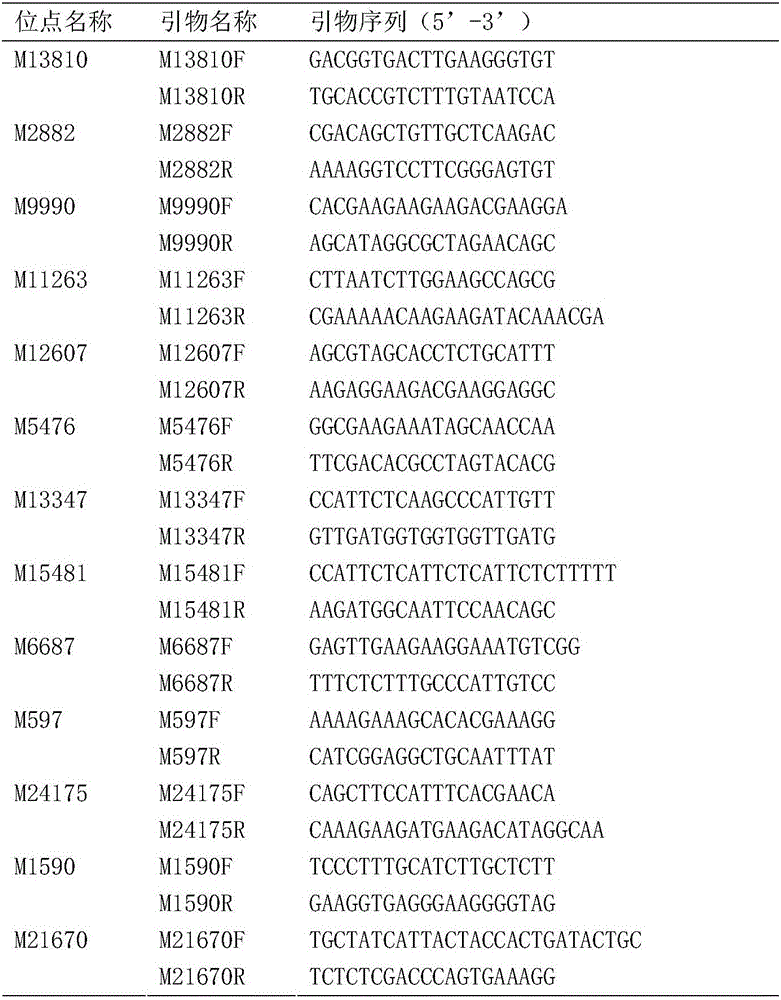
[0029] 3. Dilute the DNA extracted above to 50-100ng / μl respectively, use 13 pairs of high polymorphic microsatellite marker primers of the present invention to carry out PCR amplification, and the microsatellite pr...
Embodiment 2
[0037] Example 2: Application of the method for molecular identification of Litopenaeus vannamei germplasm
[0038] In view of the fact that the method established in the present invention has a high identification accuracy rate for different germplasms, the method of the present invention can be used to realize the identification of different germplasms of Litopenaeus vannamei and the species traceability of unknown source materials. The options offered are as follows:
[0039] 1. Collect germplasm materials from different domestic and international commercial germplasm companies. The number of individuals of each germplasm source is more than 30, and try to include materials from multiple generations and multiple regions. The DNA of the above-mentioned materials was extracted using the Tiangen Plant Genome Extraction Kit. For the method, refer to the kit instruction manual. The concentration of the extracted DNA was measured using a Nanodrop 1000 (Thermo, USA), and stored in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com