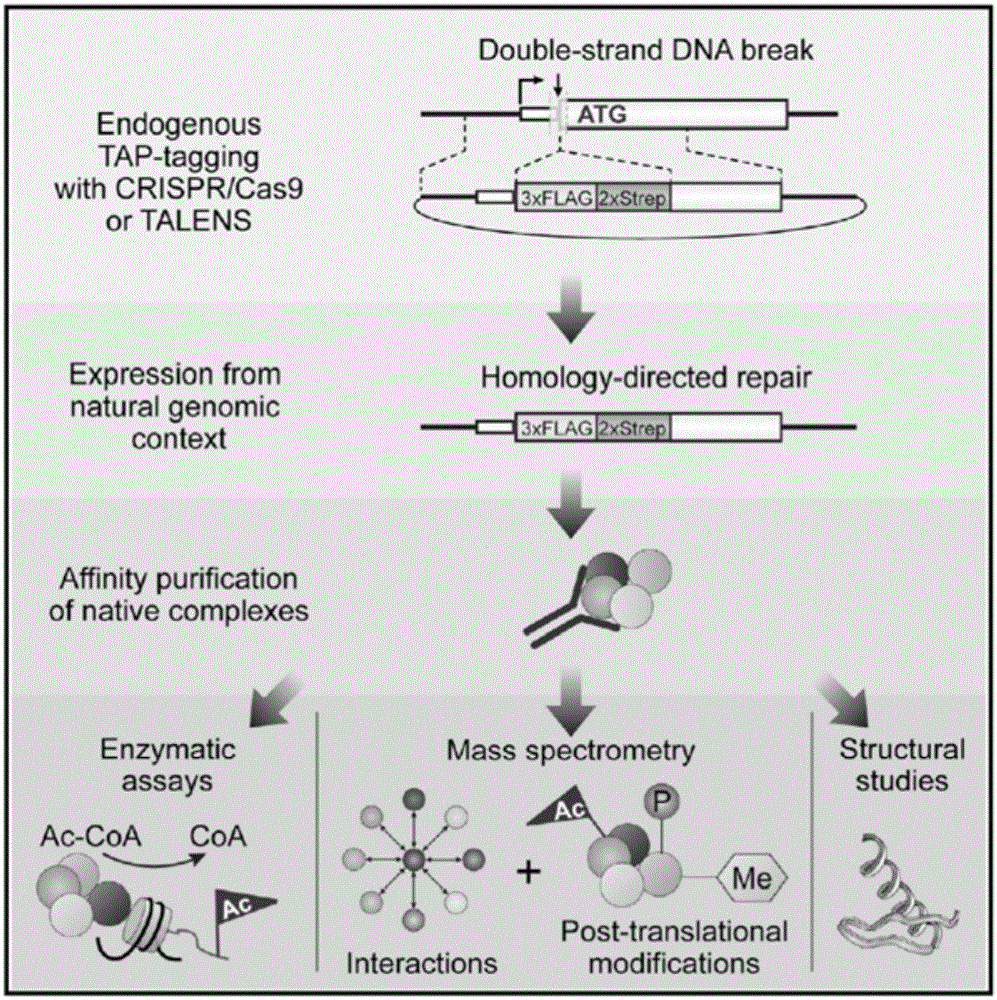
Endogenous protein marking method used for Chip-seq genome-wide binding spectrum
A genome-wide, protein labeling technology, applied in the field of protein labeling, can solve the problem of unclear transcriptional regulation mechanism of transcription factors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Marking of the NCOA1 gene in breast cancer cell MCF7 and its genomic transcriptional profiling. Concrete method based on the present invention is as follows:
[0037] (1) sgRNA library establishment:
[0038] Use the online design website http: / / cistrome.org / crispr / tool developed by our laboratory to design sgRNA for the upstream and downstream 50bp region of the stop codon site (stop codon site) of the three ERα co-transcription factor genes, and introduce the DNA open To further screen the sgRNA located on the genomic DNA with a high degree of openness.
[0039] (2) Construction of a lentiviral vector containing sgRNA:
[0040] The sequence 5'-GACCA-3' was added to the 5' end of the sgRNA sequence, and the sequence 5'-GACCA-3' was added to the 5' end of the reverse complementary sequence of the sgRNA. Synthesize modified sgRNA sequences. The lentiviral plasmid is the lentiCRISPRv2 plasmid with Cas9 and Puro elements, such as Figure 4 shown. The lentiCRISPRv2 ...
Embodiment 2
[0054] CRISPR / Cas9 applied to the liver cancer cell line HepG2 is applied to a CRISPR / Cas9 enrichment sequencing method for large-scale screening of cancer genes, including the following steps:
[0055] (1) Design sgRNA:
[0056] Use the online design website http: / / cistrome.org / crispr / tool developed by our laboratory to design sgRNA for the upstream and downstream 50bp region of the stop codon site (stop codon site) of the three ERα co-transcription factor genes, and introduce the DNA open To further screen the sgRNA located on the genomic DNA with a high degree of openness.
[0057] (2) Construction of a lentiviral vector containing sgRNA:
[0058] The sequence 5'-GACCA-3' was added to the 5' end of the sgRNA sequence, and the sequence 5'-GACCA-3' was added to the 5' end of the reverse complementary sequence of the sgRNA. Synthesize modified sgRNA sequences. The lentiCRISPRv2 plasmid with Cas9 and Puro elements was selected as the lentiviral plasmid. The lentiCRISPRv2 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com