Method for solving allele amplification unbalance by PCR additive
An allele amplification and allele technology, which is applied in the field of unbalanced allelic amplification and can solve problems such as unbalanced allelic amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0026] Example, Resolving Allelic Amplification Imbalances by PCR Additives
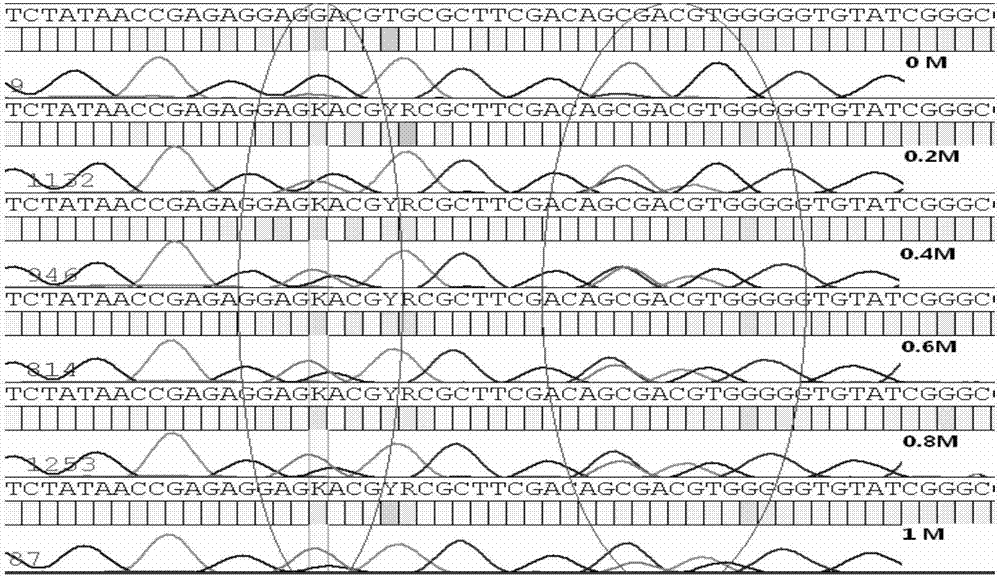
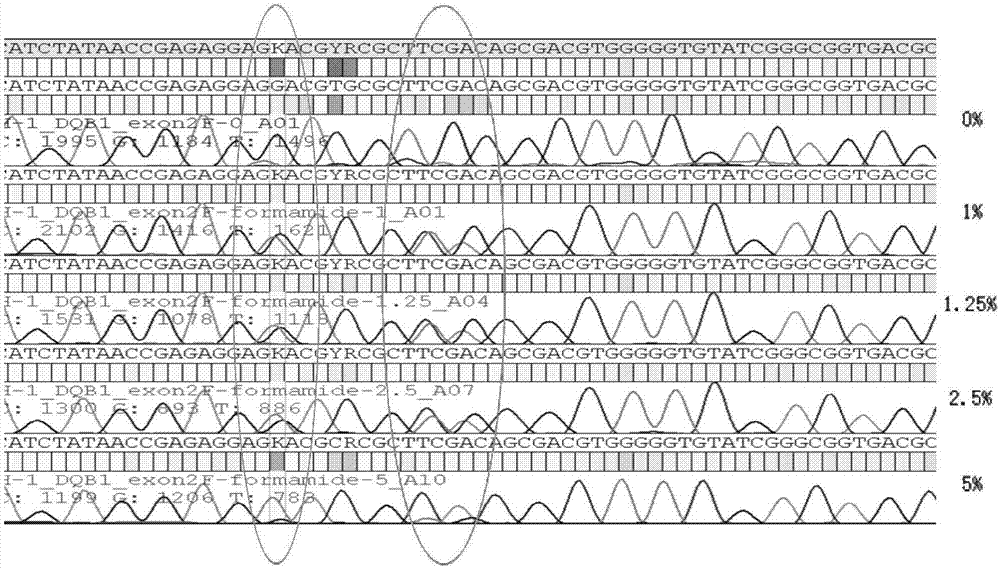
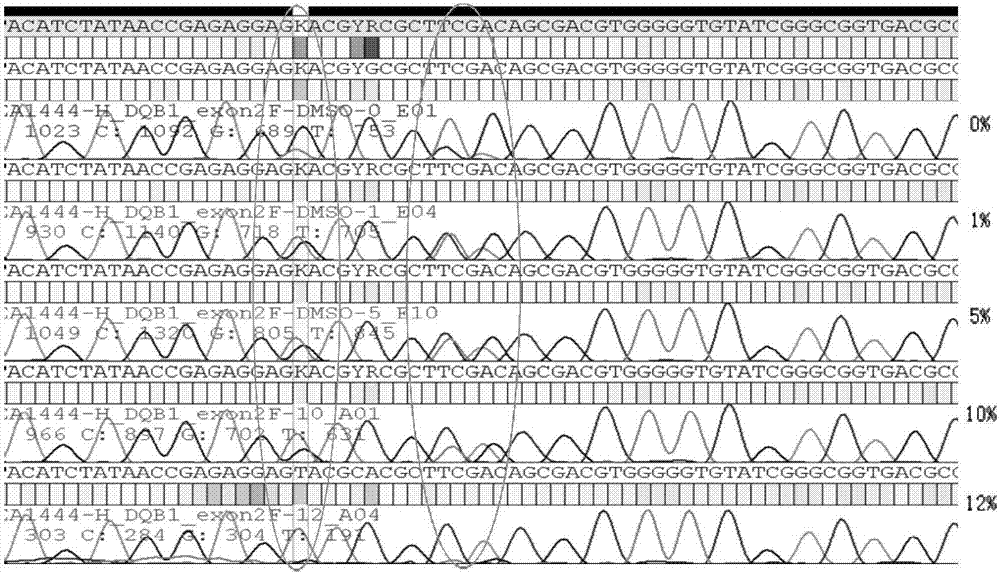
[0027] PCR additive is a kind of synergist in PCR reaction, common types include formamide, betaine, DMSO, etc. The present invention finds that the PCR additive can reduce the stability of the double-helix DNA, make the secondary structure easy to open, and help the DNA polymerase to extend through the secondary structure region. During the PCR process, the single strand of one allele of the two alleles forms a complex secondary structure or the DNA double strands melt insufficiently, resulting in inconsistent amplification efficiencies of the two alleles, that is, the problem of unbalanced amplification , You can add a certain concentration of betaine-like PCR additives to solve such problems.
[0028] The method for solving the problem of unbalanced allelic amplification by using PCR additives will be described below with specific examples.
[0029] Alleles: HLA-DQB1 genes whose genotypes are DQ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com