A Method for Protein Conformation Space Optimization Based on Parsimonious Abstract Convex Estimation
An optimization method, protein technology, applied in proteomics, used to analyze two-dimensional or three-dimensional molecular structure, genomics, etc., can solve the problems of low conformational search efficiency, high calculation cost, slow convergence speed, etc., and achieve a balanced population The effect of diversity and sampling efficiency, reducing computational cost, and reducing the number of evaluations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0038] The present invention will be further described below in conjunction with the accompanying drawings.
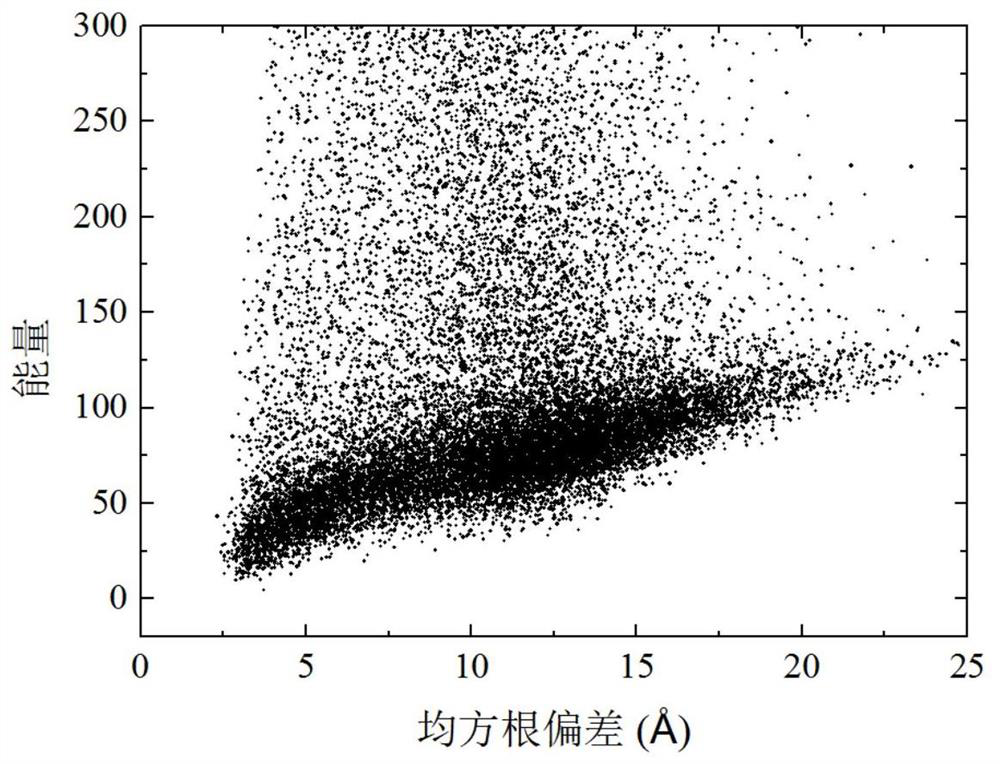
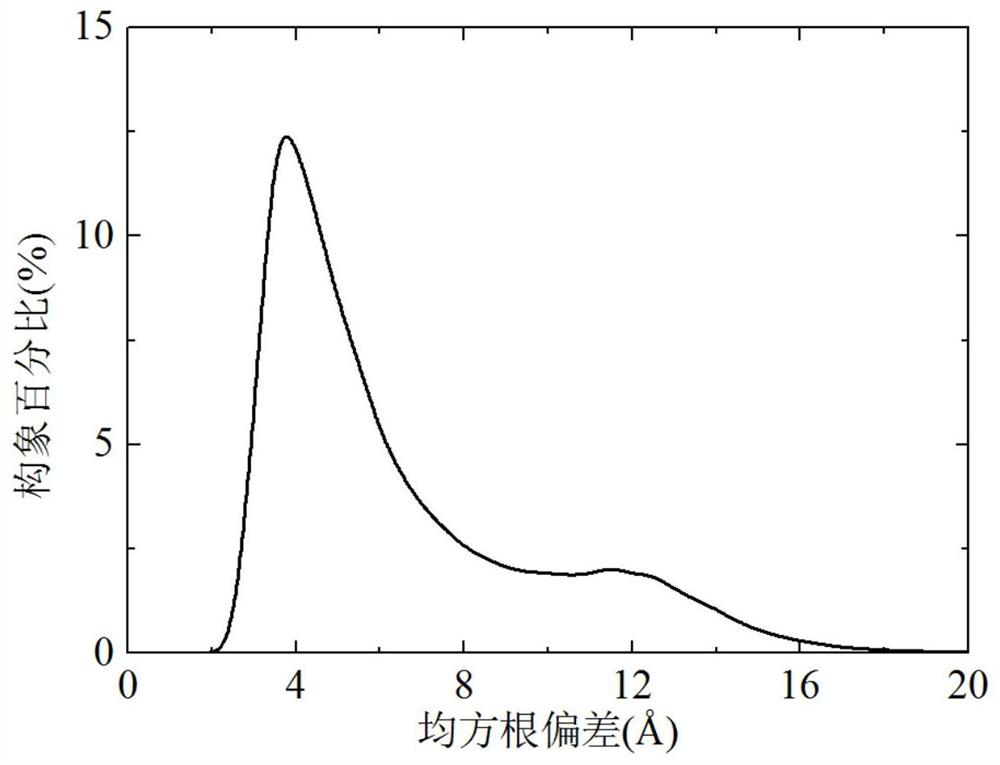
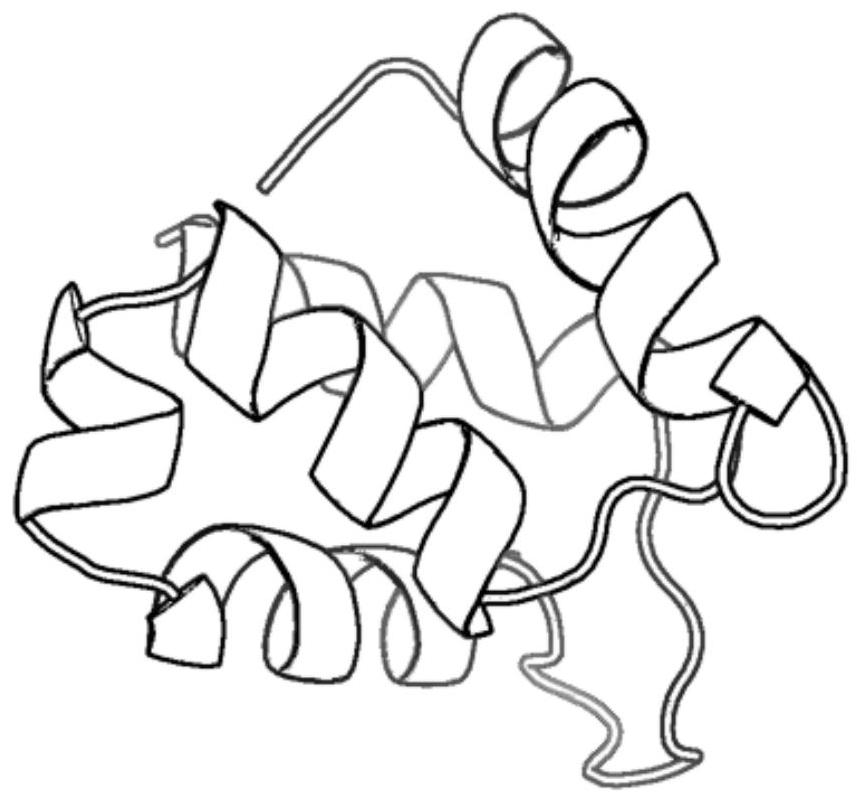
[0039] refer to Figure 1 ~ Figure 3 , a protein conformation space optimization method based on parsimony abstract convex estimation, including the following steps:
[0040] 1) Input the sequence information of the protein to be tested;
[0041] 2) Obtain the fragment library from the ROBETTA server (http: / / www.robetta.org / ) according to the sequence information;
[0042] 3) Parameter setting: set the population size NP, the crossover probability CR, the fragment length l, and the maximum number of iterations G max , learning algebra G len , the slope control factor M, and initialize the number of iterations G=0;
[0043] 4) Population initialization: Randomly assemble the fragments corresponding to each residue to generate an initial population P={C 1 ,C 2 ,...,C NP}, C i ,i={1,2,…,NP} is the i-th conformational individual in population P;
[0044] 5) For ea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com