Primer combinations, methods and kits for constructing multiple hemophilia targeting libraries based on high-throughput sequencing
A primer combination and construction method technology, applied in biochemical equipment and methods, combinatorial chemistry, chemical library, etc., can solve the problems of gene coverage, random missed detection, cumbersome operation, etc., to avoid workload and economic costs, save The effect of time cost and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Primer combination
[0025] A total of 128 target regions were targeted at all exons, splicing regions, partial promoter regions and intron regions of 7 genes including F8, F9, F11, vWF, F2, F7 and SRY. Among them, the F8 and F9 genes also include the promoter region, the F2 gene also includes a part of the 3'UTR region, and the F8 gene also includes a part of intron 4. A total of 225 pairs of primers were designed and synthesized, and their base sequences are shown in SEQID No. 1~shown in SEQ ID No.450.
[0026] The detected genes and coverage areas are as follows:
[0027]
[0028] The optimized primers are as follows:
[0029]
[0030]
[0031]
[0032] Send the above-mentioned SEQ ID No.1 to SEQ ID No.450 to Biological Company for synthesis, and mix all or part of the primer pairs of SEQ ID No.1 to SEQ ID No.450 in equal amounts, and load them in the same packaging tube or at most 2 packages In the tube, a pool of primers is formed, which ca...
Embodiment 2
[0033] Example 2 Detection sample processing and DNA extraction
[0034] The detection sample of the present invention may be whole blood, blood clot, fresh pathological tissue, paraffin-embedded tissue, and this embodiment only uses whole blood sample as an example for illustration.
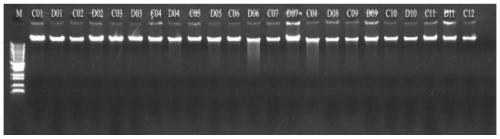
[0035] In order to reduce the interference of various anticoagulants on the PCR reaction, venous blood should be collected with EDTA-K2 anticoagulant blood collection tubes. The venous blood should be extracted and purified every day. If it cannot be extracted in time on the same day, it should be stored in a 4°C refrigerator. save. Conventional DNA extraction and purification methods such as DNA extraction kits or automatic DNA extractors can be used. The DNA concentration was detected with a Nanodrop 2000 trace nucleic acid and protein detector, and the 260 / 280 ratio and 260 / 230 ratio were determined. The extracted DNA is subjected to 0.5% agarose gel electrophoresis, and a clear electrophor...
Embodiment 3
[0036] Example 3 Constructing Multiple Hemophilia Targeting Libraries
[0037] Standardize the sample DNA extracted in Example 2, adjust the concentration to 40-50ng / ul as the amplification template, and use all or part of the primers from SEQ ID No.1 to SEQ ID No.450 described in Example 1 The primer mixture pool composed of primers is used for ultra-high multiplex PCR amplification. Perform PCR amplification according to the following amplification system and conditions, and perform ultra-high multiplex PCR amplification according to the multiple ratio volume or equal concentration system of the following system:
[0038]
[0039] Preferably, the amplification conditions are: 95°C for 10 min; 98°C for 15 s, 60°C for 5 min, 9-11 cycles; 10°C ∞.
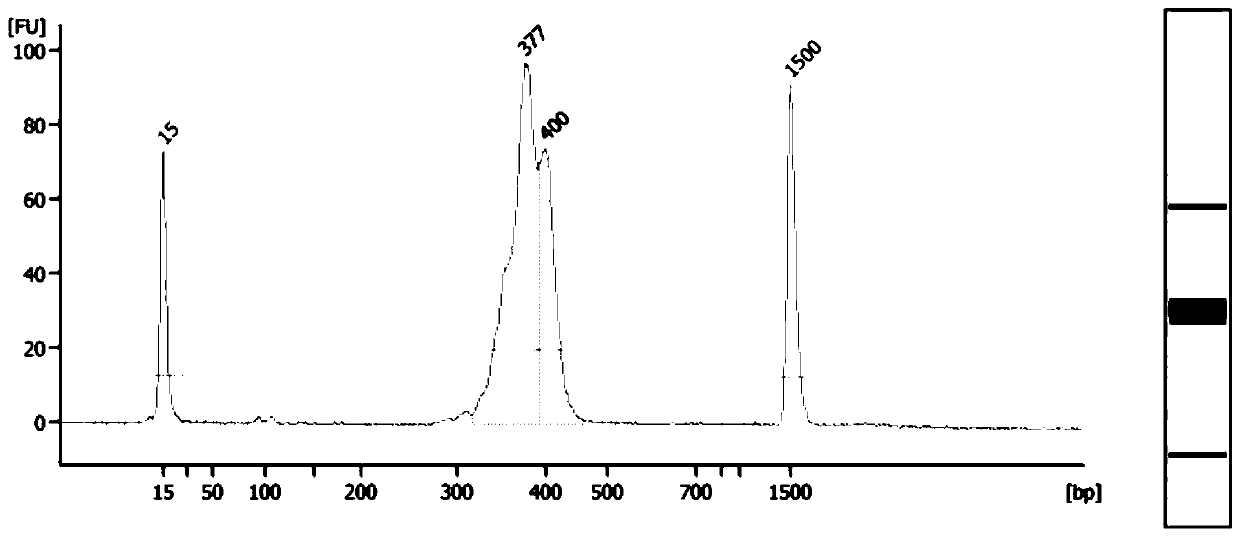
[0040] The amplification product is purified by magnetic beads, non-specific products are removed, and then purified to obtain a targeted library of the target gene. High-throughput sequencing primers and barcode tags can be add...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com