Detecting hepatocellular carcinoma
A methylation, sample technology, applied in the field of detection of hepatocellular carcinoma
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment I
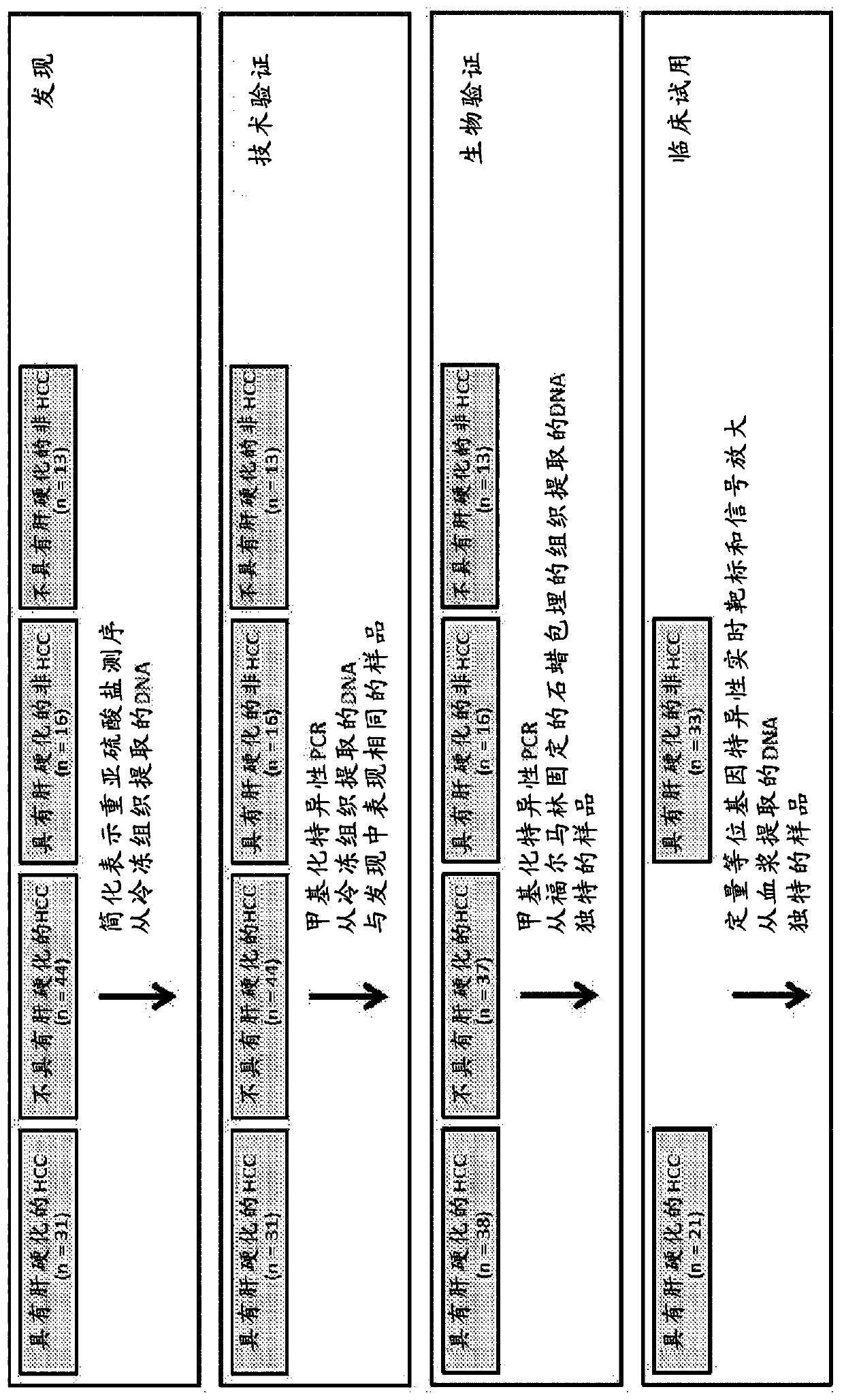
[0205] This example describes the identification of 311 differentially methylated regions (DMRs) that were used to discriminate HCC DNA samples from DNA derived from normal controls (eg, non-HCC individuals with or without cirrhosis).
[0206] Experiments were performed in four phases.
[0207] First, DNA methylation was performed using RRBS on DNA extracted from frozen tumor HCC tissue (with and without cirrhosis) and from frozen normal liver tissue (with and without cirrhosis) as well as buffy coat samples from healthy volunteers. Kylation marker discovery. Discriminatory differentially methylated regions (DMRs) were identified by stringent filtering criteria and the same or extended samples were re-analyzed using real-time methylation-specific quantitative PCR analysis (qMSP) to ensure reproducibility of results (technical validation).
[0208] Second, candidate markers were selected by blinded qMSP analysis of DNA extracted from independent archival case and control tissu...
Embodiment II
[0265]Tissue samples (75 HCC, 20 cirrhotic, and 30 normal) were run in QuARTs format on a real-time PCR instrument (Roche LC480) (see US Patent No. 8,361,720) using primers and probes generated from DMR sequences (see Table 5), GoTaq DNA polymerase (Promega), Lyase 2.0 (Hologic) and fluorescence resonance energy transfer reporter kit (FRET) (Biosearch Technologies) containing FAM, HEX and Quasar 670 dyes. Table 6 shows the ability of each marker to discriminate HCC from cirrhotic and normal samples with 100% sensitivity.
[0266] table 5.
[0267]
[0268]
[0269] Table 6.
[0270]
Embodiment III
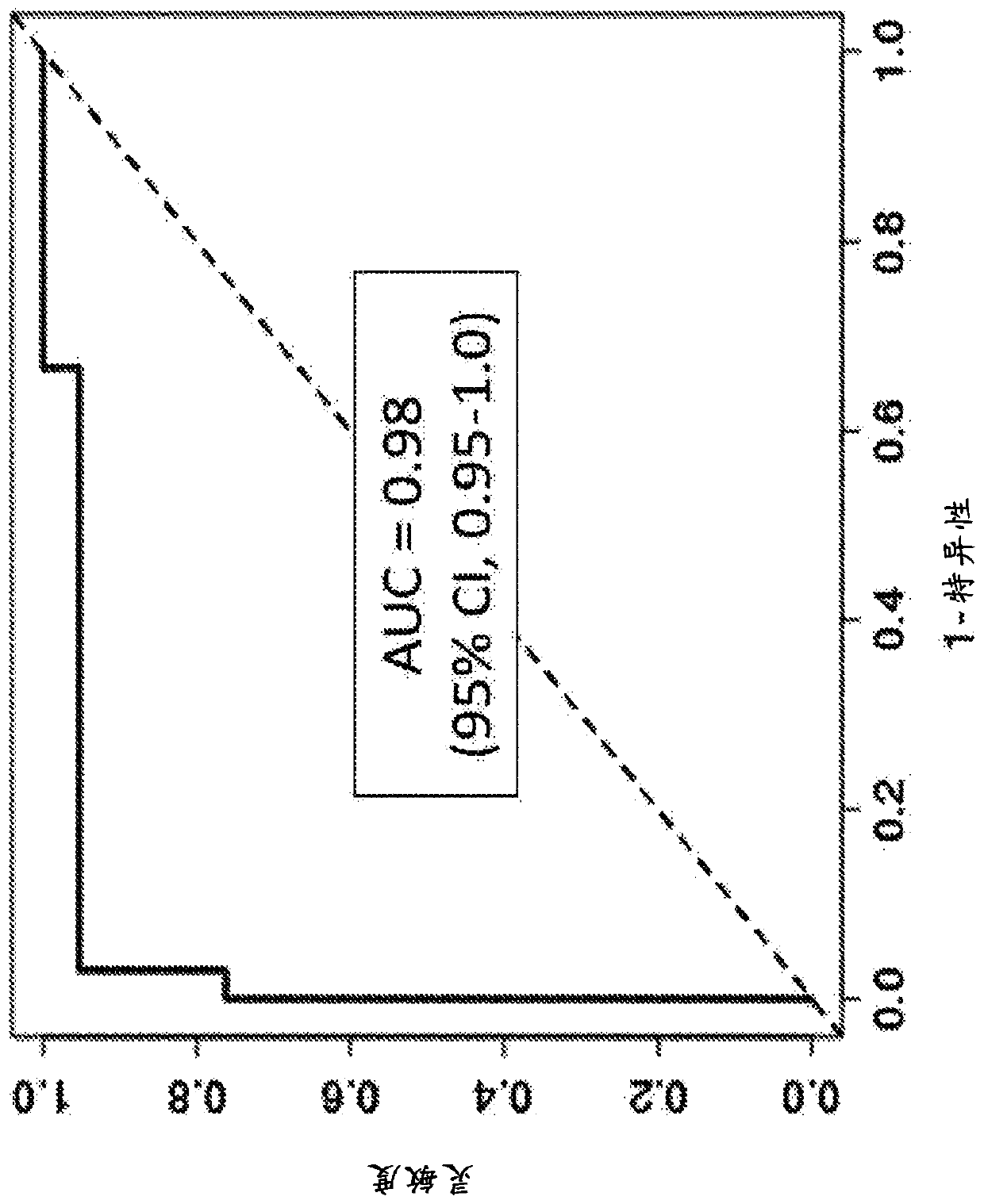
[0272] The main objective of this example is to identify a panel of markers predictive of hepatocellular carcinoma (HCC). Plasma from 244 subjects (95 HCC and 149 controls) was adjusted to 2 mL and extracted. The 149 controls consisted of 51 cirrhotic patients and 98 normal patients.
[0273] Figure 9 The relative importance of each of the methylation markers considered in this analysis is shown. The cross-validated estimates for sensitivity and specificity for the entire marker panel were 75% and 96%, respectively.
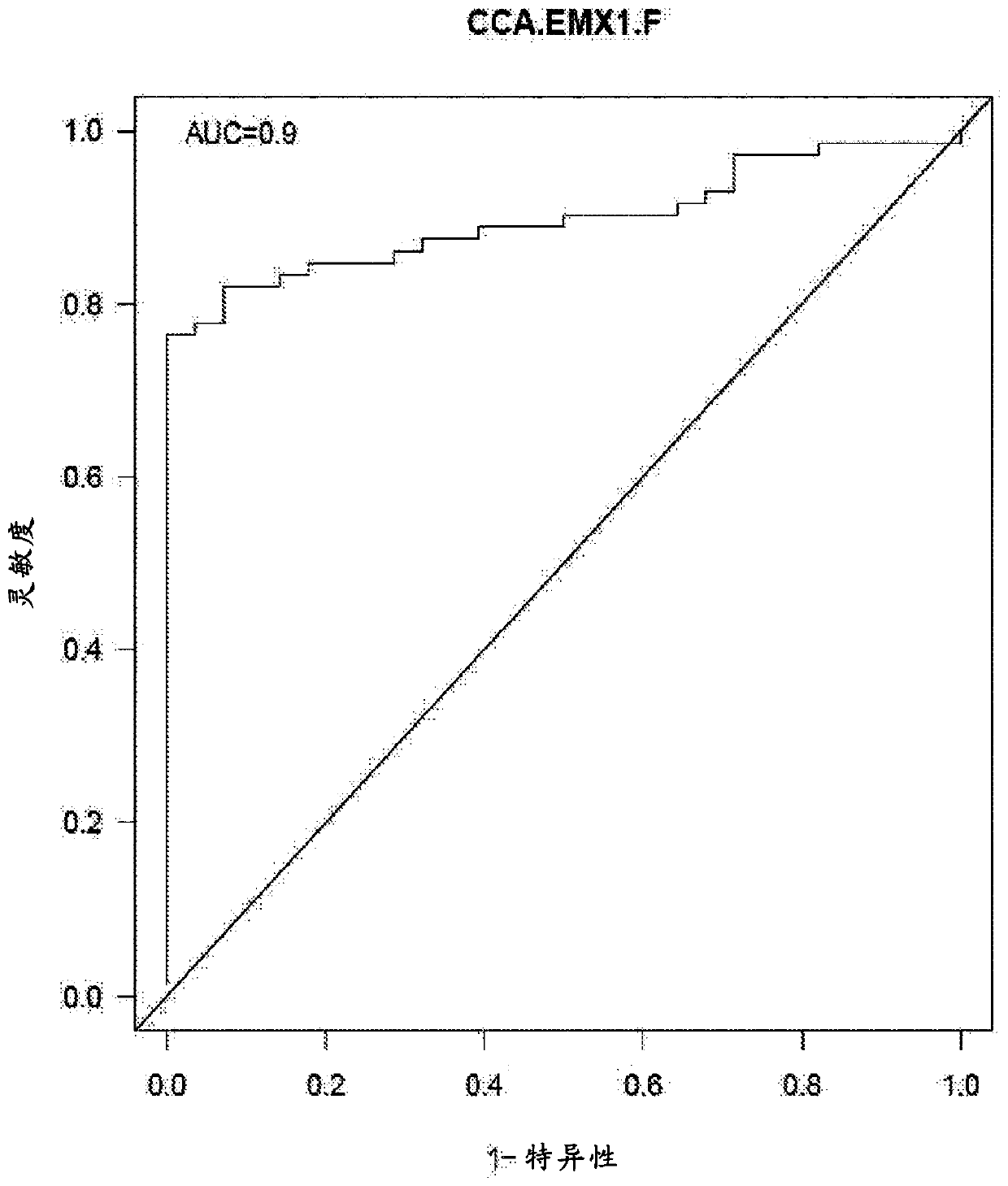
[0274] The following marker panel (Chr12.133, CLEC11A, EMX1, HOXA1, CCNJ_3707) yielded a sensitivity of 85.3% for HCC at a specificity of 88.6% in controls versus normal patients (Table 7).
[0275] Table 7.
[0276]
[0277] The same group (Chr12.133, CLEC11A, EMX1, HOXA1, CCNJ_3707) produced the following specific reductions for normal and cirrhotic (control group) patients (Table 8).
[0278] Table 8.
[0279]
[0280] An exemplary procedure for i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com