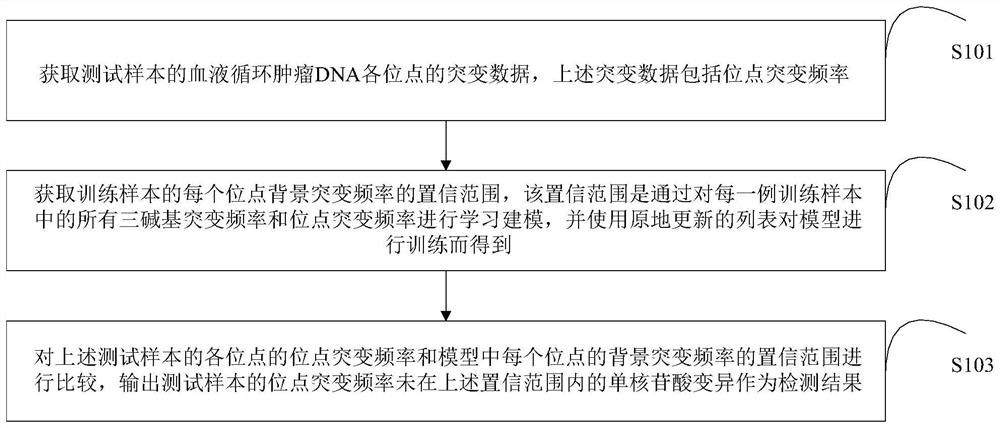
Single nucleotide variation detection method, device and storage medium based on circulating tumor DNA
A single nucleotide variation and detection method technology, applied in genomics, proteomics, instruments, etc., can solve the problems that iDES and TNER cannot be effectively detected, cannot effectively distinguish mutation frequency, and reduce the sensitivity of variation detection. Achieve the effects of reducing detection time, increasing secondary screening, and reducing false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
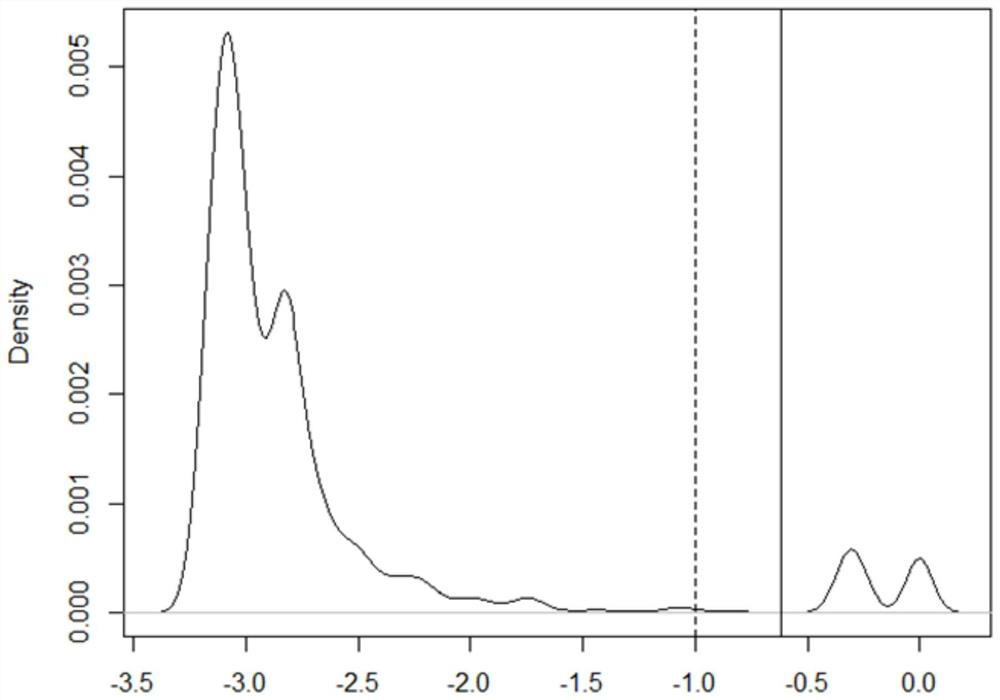
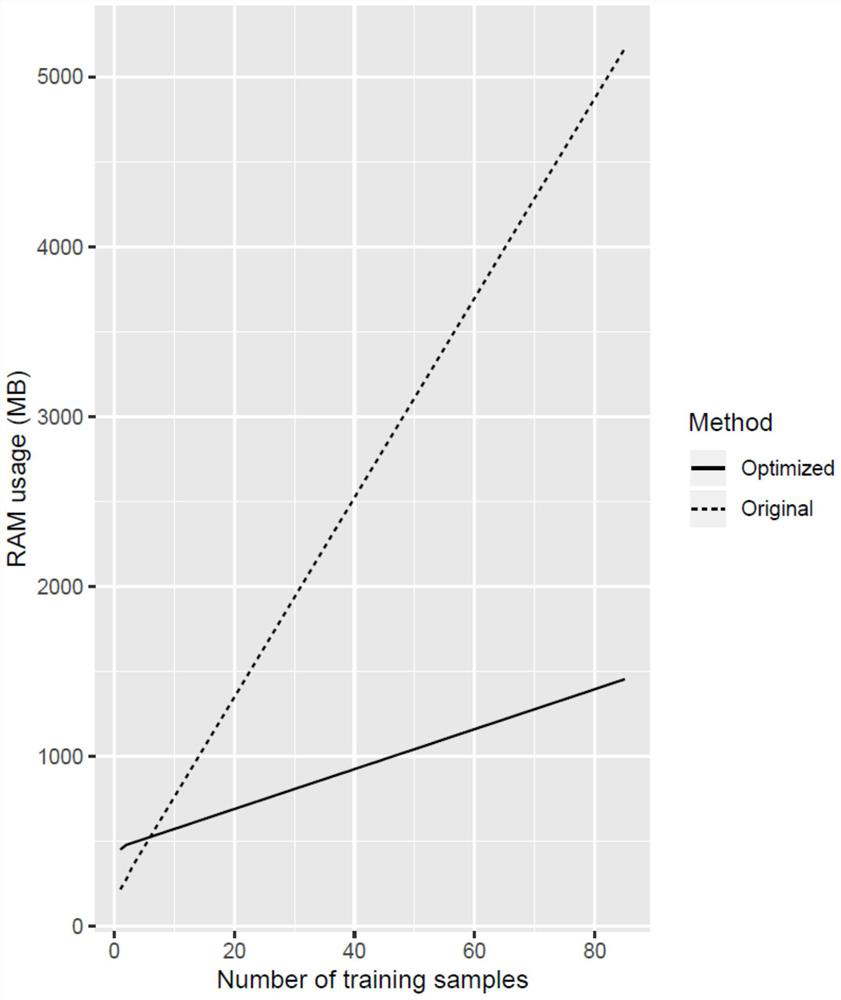
Image
Examples
Embodiment 1
[0090] In this example, training data: ACGT files generated by 10 blood samples from healthy people through the Target chip sequencing of Shenzhen Yuce Biotechnology Co., Ltd.; test data: 189 blood samples from cancer patients passed through The ACGT file generated by the targeted (Target) chip sequencing of Co., Ltd.
[0091] The blood samples from 10 healthy people were sequenced by the Target chip of Shenzhen Yuce Biotechnology Co., Ltd. and the genome sequence fragments were compared with the human reference genome, and the comparison results in BAM format were obtained. Then use Samtools software to convert the BAM format file into a pileup format file. During the conversion process, only reads with a sequencing error and alignment error rate of less than 0.1% are allowed, and the corresponding Phread Score (Phread Score) and Mapping Score (MappingScore) are both 30. Then convert the generated pileup format file into single nucleotide mutation frequency data file ACGT fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com