Nucleotide unit point variation detecting method based on neural network
A mutation detection and neural network technology, applied in the field of neural networks, can solve the problems of inaccurate detection position, low tumor purity, and reduced depth of mutation sites, and achieve the effects of low time complexity, accurate test results, and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0034] In order to make the object, technical solution and advantages of the present invention more clear, the present invention will be further described in detail below in conjunction with the examples. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
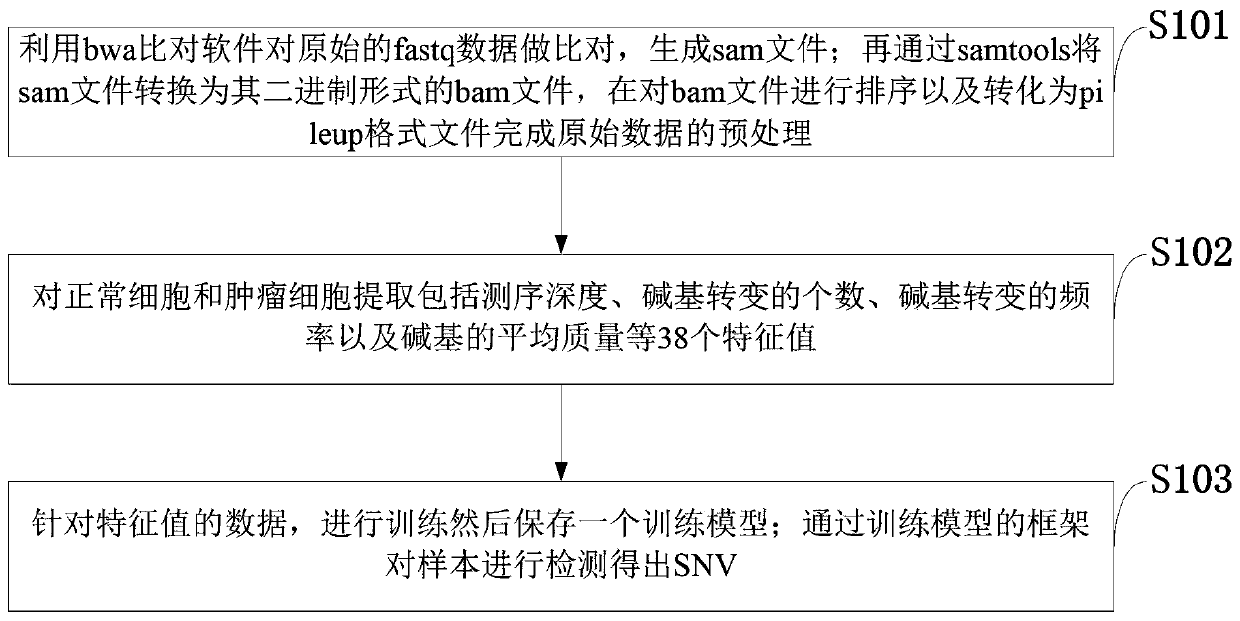
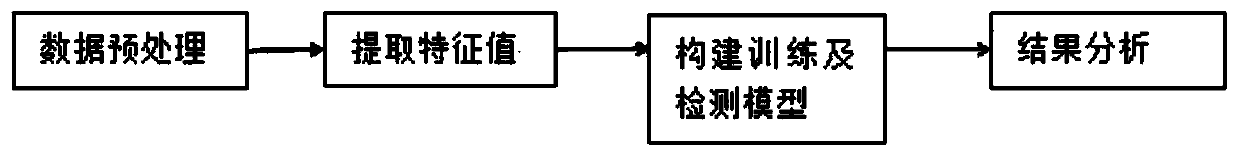
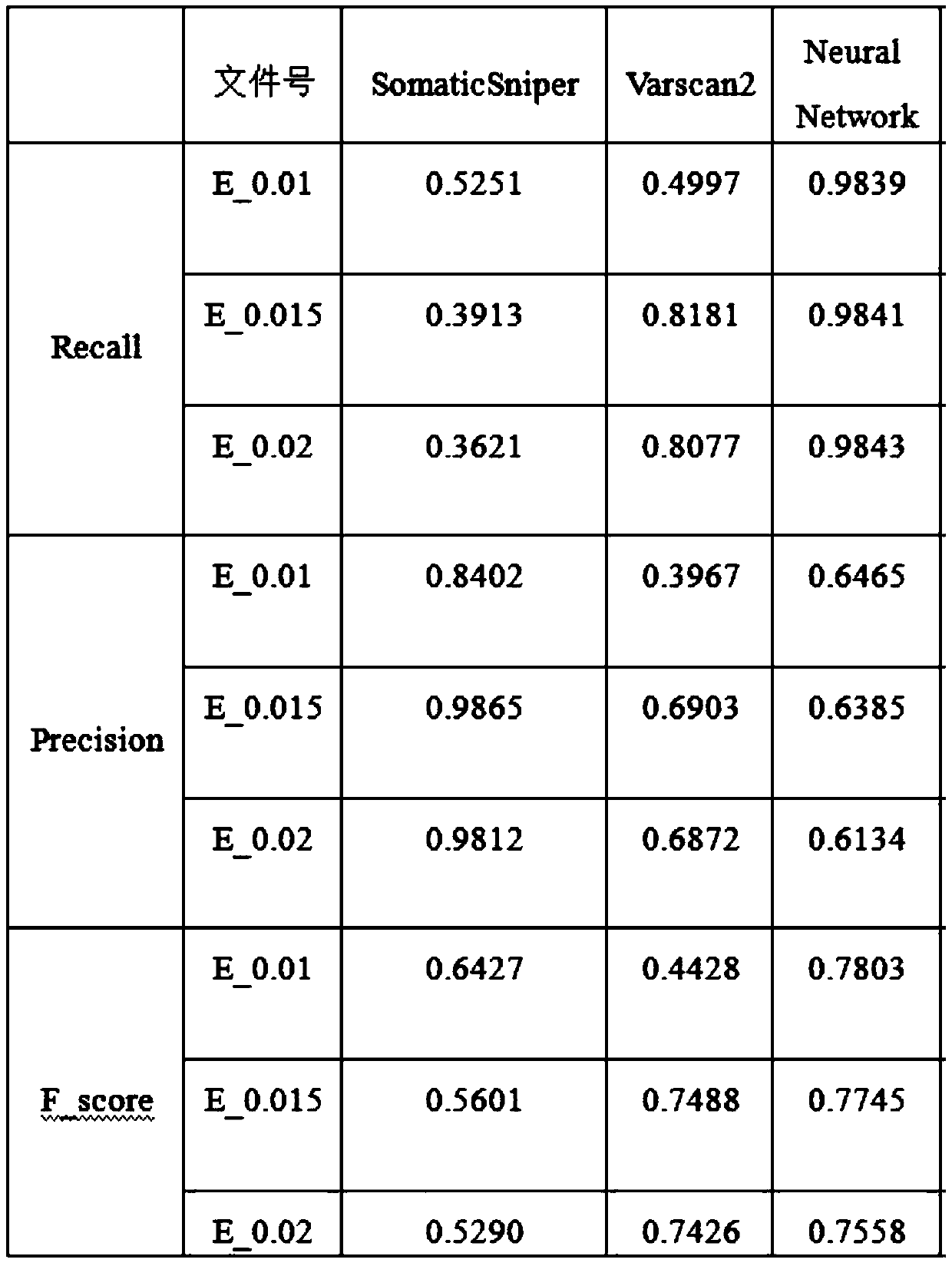
[0035] For the existing methods for detecting SNV mutations on the genome, the detection position is not accurate enough; the accuracy rate is low under low tumor purity; relying on only one read for comparison is likely to cause the omission of mutation detection. The present invention uses the eigenvalues set by genetic information; trains these eigenvalues through a TensorFlow framework to obtain a data model, and detects and screens samples according to the trained model to obtain SNV.
[0036] The application principle of the present invention will be described in detail below in conjunction with the accompanying d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com