A lncRNA-protein Association Prediction Method Based on Projected Neighborhood Nonnegative Matrix Factorization
A non-negative matrix decomposition and protein technology, which is applied in the field of lncRNA protein association prediction based on projection neighborhood non-negative matrix decomposition, which can solve the problem of low prediction accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
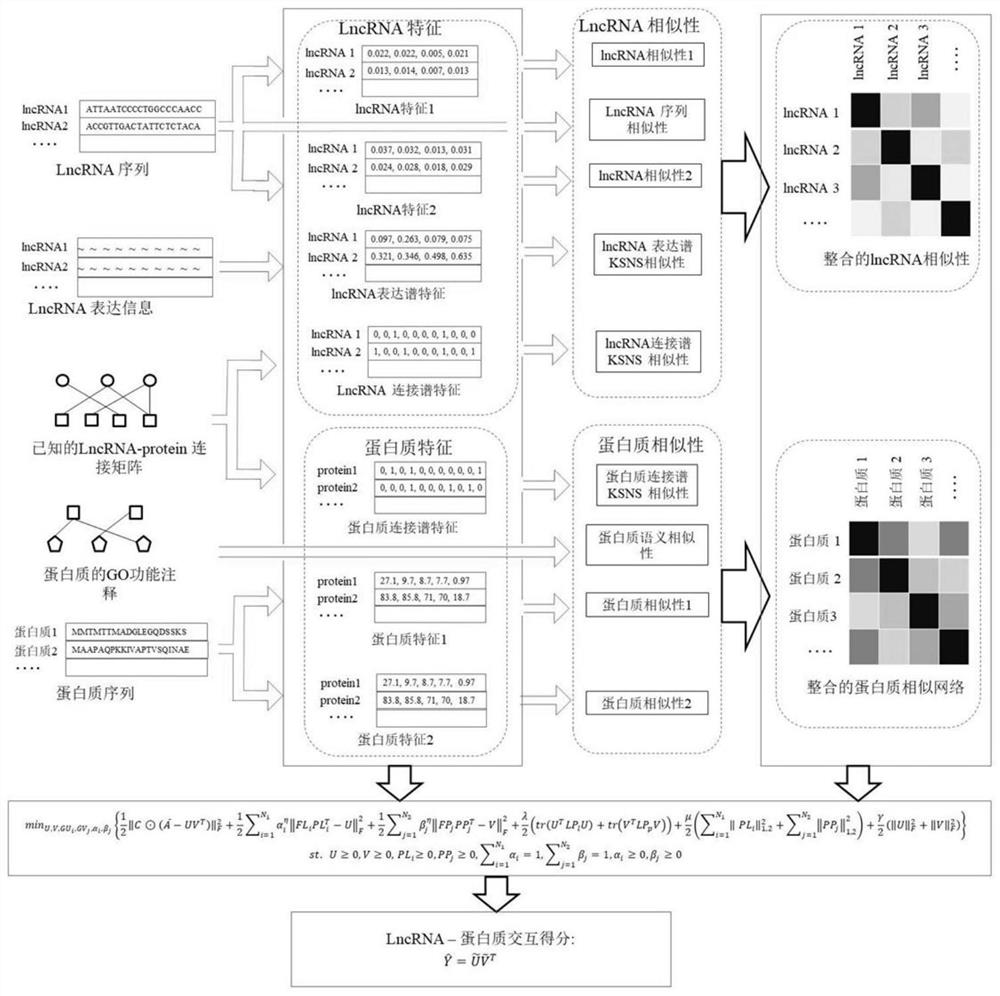
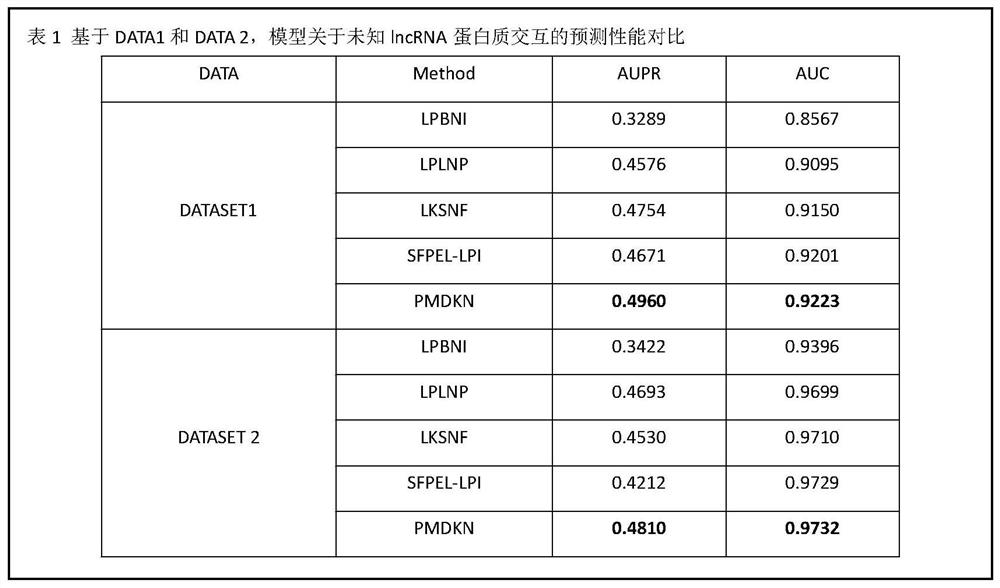
[0053] The purpose of the present invention is to provide a lncRNA-protein association prediction method based on projected neighborhood non-negative matrix decomposition for the technical problems that the prediction accuracy of the method in the prior art is not high and the interaction between unknown lncRNA and protein cannot be predicted, and achieve Improve prediction accuracy and infer the purpose of unknown lncRNA and protein interaction.
[0054] In order to achieve the above object, the main idea of the present invention is as follows:
[0055] According to the multiple features of lncRNA, multiple features of protein, lncRNA similarity matrix, protein similarity matrix, known lncRNA and protein association matrix, project lncRNA and protein to a potential common feature subspace, and then calculate The correlation between lncRNA and protein, using these correlations to prioritize, and then predict the connection between lncRNA and protein.
[0056] The invention ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com