InDel marker for identifying red/non-red properties of pulp of peaches, primer pair and application thereof
A primer pair and labeling technology, applied in the biological field, can solve the problems of cumbersome identification steps, red pulp, low accuracy, etc., and achieve low-cost effects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1, the acquisition of indel marker (InDel)
[0027] In the present invention, two germplasms of peach germplasm resources "Tianjin Shuimi (red meat)" and "Baifeng (white meat)" are used as samples of the peach germplasm resources of Zhengzhou Institute of Fruit Trees, Chinese Academy of Agricultural Sciences. Two germplasms were resequenced to obtain 18Gb data, with an average sequencing depth of about 30×. According to the reads of 10kb-100kb obtained by sequencing, compare with peach reference genome v.2.0 (ftp: / / ftp.bioinfo.wsu.edu / species / Prunus_persica / Prunus_persica-genome.v2.0.a1 / ), identify 12692 large nucleotide indels were found, among which there was a 6647bp insertion sequence in "Tianjin Shuimi" at the promoter position of BL gene (between 707474bp and 707475bp of chromosome 5 of the peach genome). But 'Baifeng' did not. Afterwards, the upstream primers were designed according to the upstream reference sequence of the insertion site, and the do...
Embodiment 2
[0028] Embodiment 2 Utilizes the method for nucleotide insertion (InDel) marker identification peach pulp color
[0029] 1. DNA extraction
[0030] The modified CTAB method is used to extract the DNA of the sample tissue (leaf) to be tested, and the DNA sample volume is not less than 30 μl. Measure the OD value of the DNA sample at 260nm and 280nm with a UV spectrophotometer, and calculate the DNA content and OD 260 / 280 ratio. DNA sample purity OD 260 / 280 The value should be between 1.8-2.0, and the concentration should be diluted to around 10ng / μl.
[0031] 2. Primer design
[0032] Primers were designed according to the upstream reference sequence of the insertion site and the 5' end sequence of the insertion fragment (about 1680 bp in total), as shown in Table 1 below.
[0033] Table 1 Sequence information at both ends of the insertion site
[0034]
[0035]
[0036] Note: The nucleotide sequence in the gray bottom part is the reference sequence upstream of the ...
Embodiment 3
[0048] Example 3, 3 hybrid populations using the invented InDel markers to carry out performance blind test verification
[0049] 1. Selection of test materials
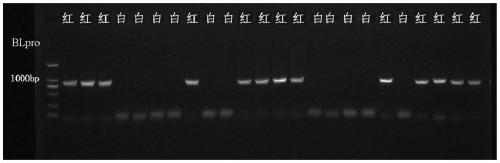
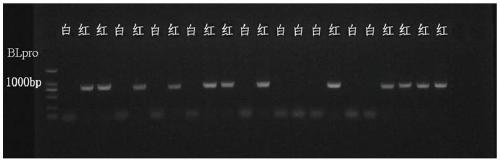
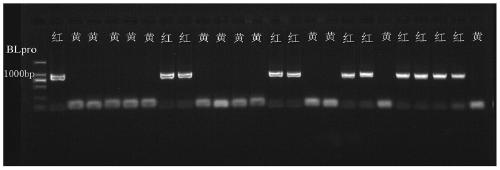
[0050] Using conventional peach varieties planted in the resource garden of Zhengzhou Fruit Tree Research Institute as experimental materials, 3 red-fleshed peach hybrid populations with investigated phenotypic traits were selected from them, with a total of 455 individual plants. See Table 5 for details.
[0051] Table 5 The name and population size information (unit: strain) of the hybrid population for testing
[0052]
[0053] Note: 07-4-28 and 09-1 Xi-28 are excellent red meat lines, Yangzhou 431 is a white meat local variety, Zhaohui is a white meat bred variety, Zhongyou 13 is a yellow meat bred variety.
[0054] 2. Utilize the identification method of InDel mark of the present invention
[0055] Using the InDel marker of the present invention, a total of 455 single-plant pulp colors of 3 red-flesh peach ...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap