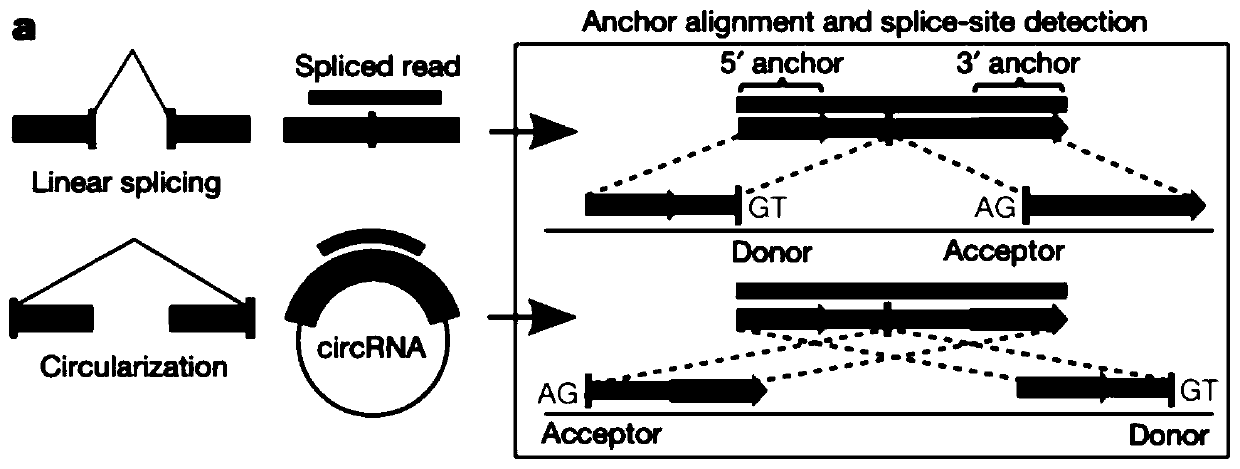
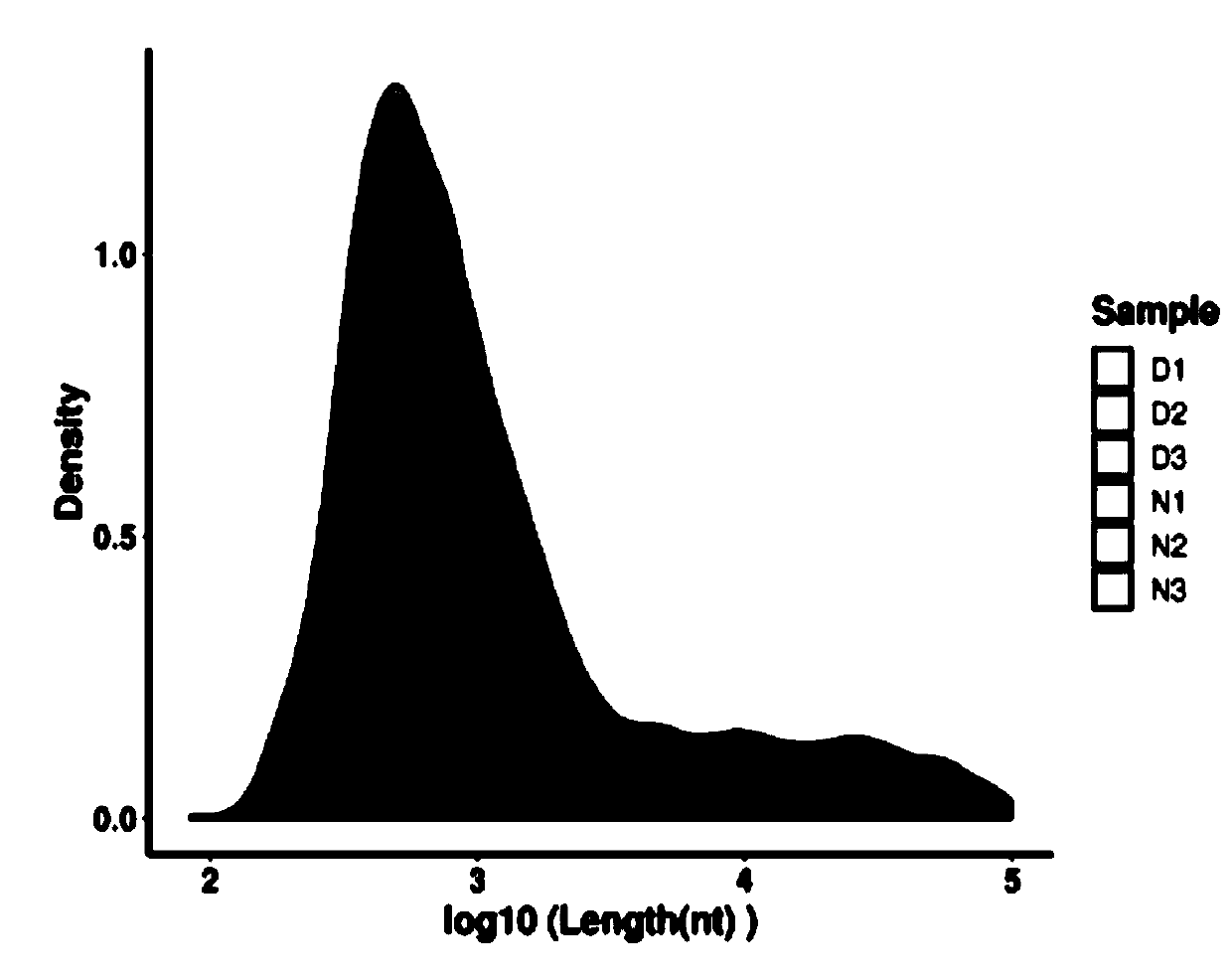
Circular RNA identification and expression quantification analysis method
An analysis method, a circular technology, applied in the field of gene detection, can solve problems such as inability to find circRNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
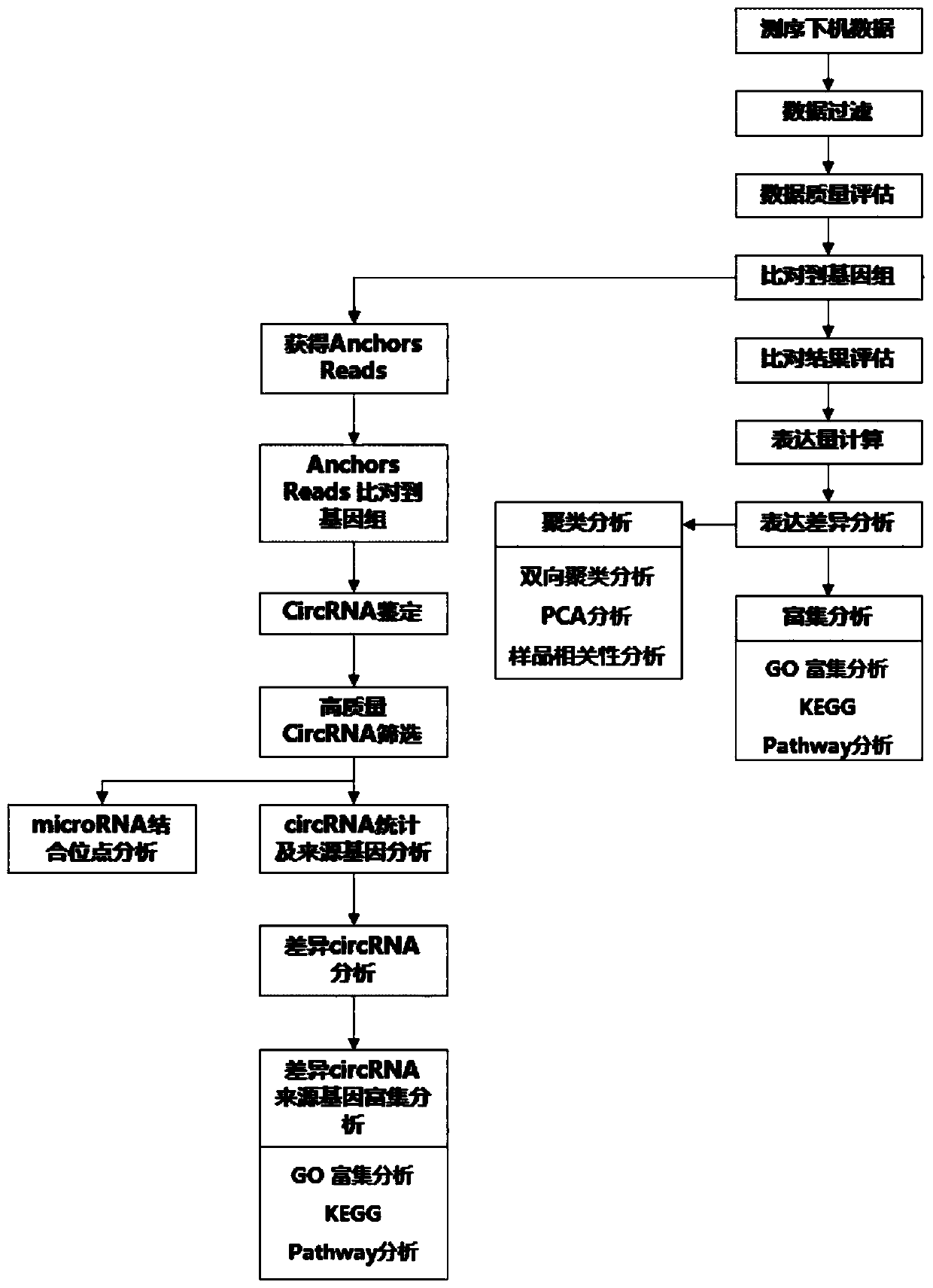
[0047] An analysis method for circular RNA identification and expression quantification, comprising the steps of:
[0048] Step 1, sequencing data filtering step:
[0049] Use fastp software to remove adapters and low-quality sequences in the sequencing results:
[0050] Fastp uses PE reads overlap information to automatically identify adapter sequences, with higher accuracy and removal efficiency. At the same time, the length of 5 bases was used as a window, and the window was slid from the 3' end to the 5' end, and the windows with an average quality less than 20 in the window were cut off. Finally, reads with a length greater than 50 are retained.
[0051] The sequencing data after removing adapters and low-quality sequences were quality-controlled using fastqc software:
[0052] According to the sequencing characteristics of RNASeq, the four lines of the base distribution of the sequencing data should be: AT parallel and close, GC parallel and close; the overall distrib...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com