Drug virtual screening system and method for crystal complex
A technology of crystal complexes and virtual screening, applied in biostatistics, bioinformatics, chemostatistics, etc., can solve problems such as difficulty in obtaining new skeletons, achieve the effects of reducing intervention, improving efficiency, and ensuring safety
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
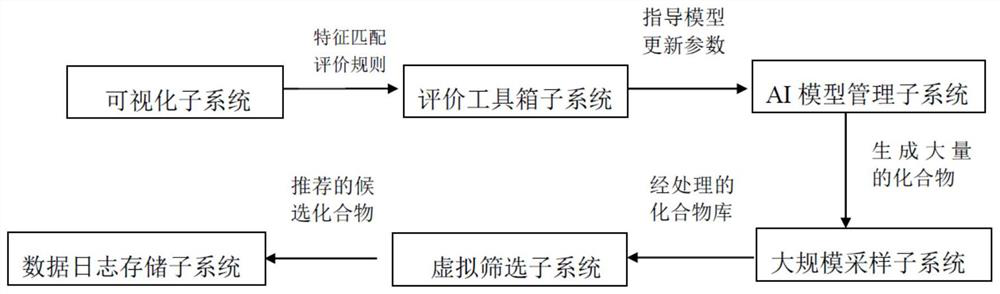
Image
Examples
Embodiment 1
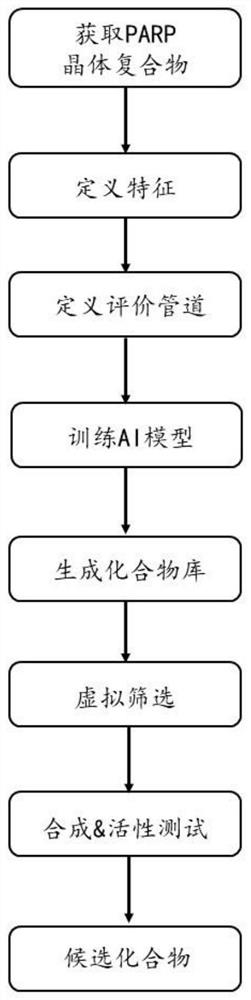
[0052] Such as figure 2 The flow shown:
[0053] Polyadenosine diphosphate-ribose polymerase (PARP) participates in base repair by catalyzing ADP ribosylation, plays an important role in the repair of single-strand DNA damage in cells, and is one of the targets of anticancer drugs. PARP1, a subtype of PARP, is one of the targets for the treatment of triple-negative breast cancer. Starting from the crystal complex of PARP1, follow the steps shown in the protocol (eg figure 2 shown) for drug design.
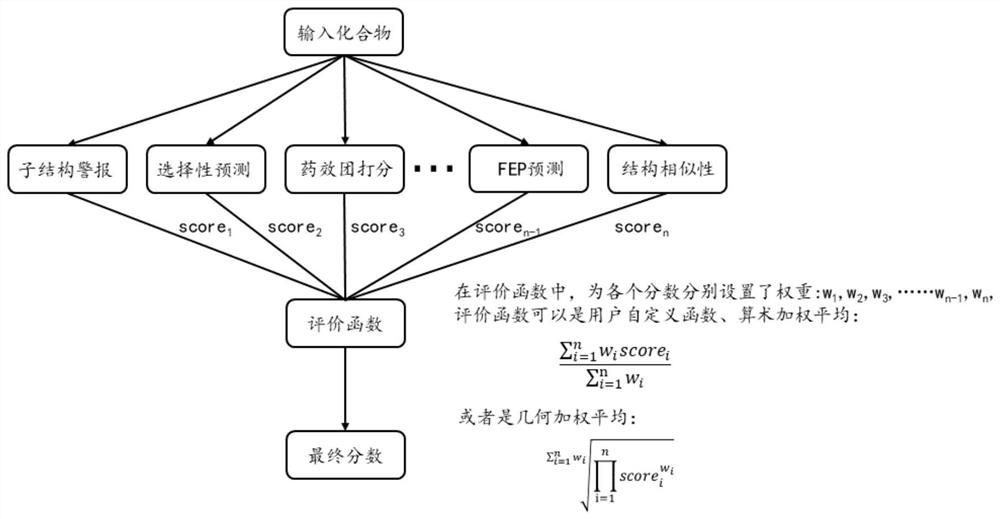
[0054] (1) Download the crystal complex structure of PARP1 from the protein crystal structure database. Through the visual analysis of the crystal complex of PARP1, combined with the binding mode reported in the literature, four key pharmacophore features (a hydrogen bond donor feature, a hydrogen bond receptor feature, and two hydrophobic features), and assign weights to the four features (the weights are 3, 3, 2, 1 in turn) to integrate into a pharmacophore feature evaluatio...
Embodiment 2
[0063] Alzheimer's disease is a representative degenerative disease of the central nervous system. Several studies on Alzheimer's disease have reported multiple targets in the literature. Acetylcholinesterase is one of the important targets. Using crystal complexes of acetylcholinesterase and its inhibitors as a starting point, search for inhibitors with novel scaffolds.
[0064] (1) According to the reports in the literature, one of the crystal complexes (PDB: 4EY7) was used as a starting point. Through the visual analysis of the crystal complex (PDB: 4EY7), combined with literature reports, the ligand was located and 5 key pharmacophore features were identified, including 2 hydrogen bond acceptors and 2 aromatic ring features , 1 hydrophobic feature, and the weight of the pharmacophore feature is 1, which is integrated into a target feature evaluation module.
[0065] (2) Combining the pharmacophore model defined in step (1) into a pharmacophore evaluation module, and sup...
Embodiment 3
[0071] Heat shock protein 90 is a new target of anti-tumor drugs discovered in recent years. Inhibitors of heat shock protein 90 can destroy the structure and degradation process of proteins in vivo to play an anti-tumor role. After the crystal structure of heat shock protein 90 was published, computer-aided drug design has become the mainstream of research and development of new heat shock protein 90 inhibitors. This example attempts to start with the crystal complex of heat shock protein 90 and recommend a batch of novel heat shock protein 90 inhibitors.
[0072] (1) One heat shock protein 90 (PDB: 1YET) was used as a starting point. Through the visual analysis of heat shock protein 90 (PDB: 1YET), combined with literature reports, the binding position of the inhibitor on heat shock protein 90 (PDB: 1YET) was defined, and 2 hydrogen bond acceptors, 2 hydrophobic centers and Two hydrogen bond donors form a pharmacophore model, and the weight of these pharmacophores is 1, whi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com