Targeted Sequencing Methods for Detecting Gene Fusions
A technology of targeted sequencing and gene fusion, applied in the field of targeting, can solve the problems of reduced NTRK3 fusion sensitivity, low throughput, and high false positive rate, so as to reduce the cost of probe synthesis and sequencing, improve detection sensitivity, and reduce the number of probes Quantity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
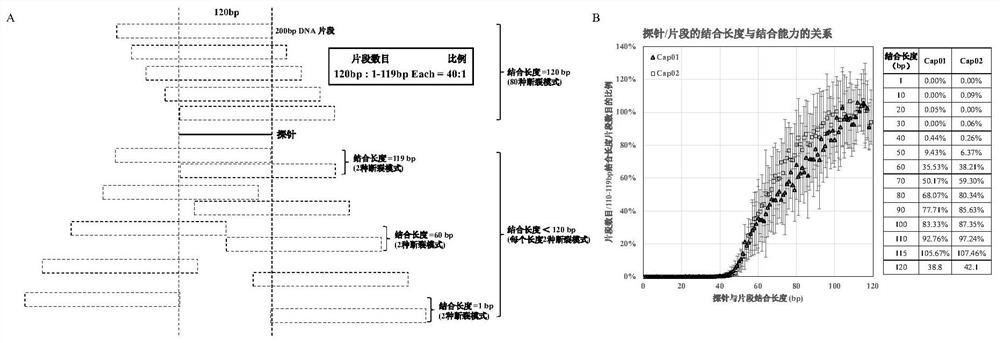
[0044] This example studies the effect of the binding length of the probe and the target fragment on the binding force of the probe.
[0045] 1. Probe selection:
[0046] In order to detect the effect of the binding length of the probe and the target fragment on the binding force of the probe, a human genome probe pool with a size of about 100Kb was used for capture and sequencing, and then no other probes within the upstream and downstream 1Kb were selected, and the GC content was 47.5-52.5 % of the 16 probes corresponding to the interval were analyzed.
[0047] Table 1
[0048]
[0049]
[0050]
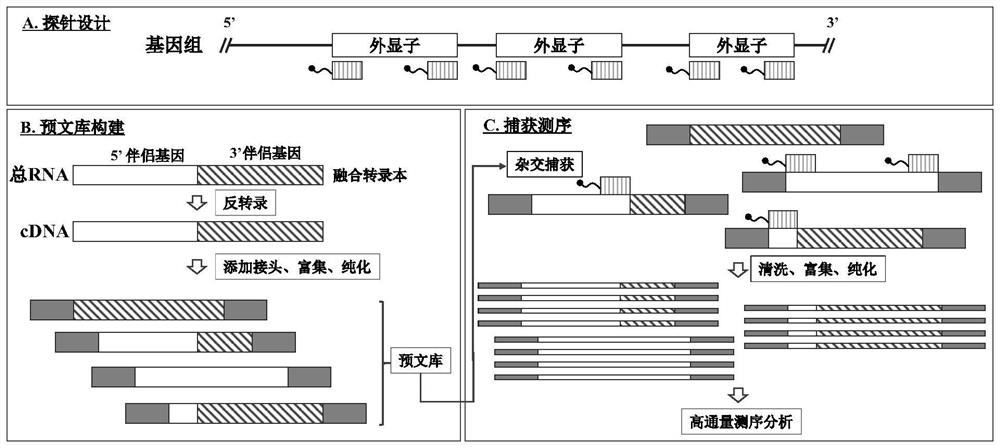
[0051] 2. Pre-library construction and capture sequencing:
[0052] A DNA library building kit (Rapid DNA Lib Prep Kit, ABclonal) was used to construct the pre-library used in this example on NA12878gDNA (Coriell) (insert size: ~200 bp; PCR cycle number: 7).
[0053] Follow the steps as indicated in A-J for 4 hours of hybrid capture.
[0054] A. Library pre-blocking
...
Embodiment 2
[0108] This embodiment is an example of fusion gene detection.
[0109] 1. Probe design and synthesis:
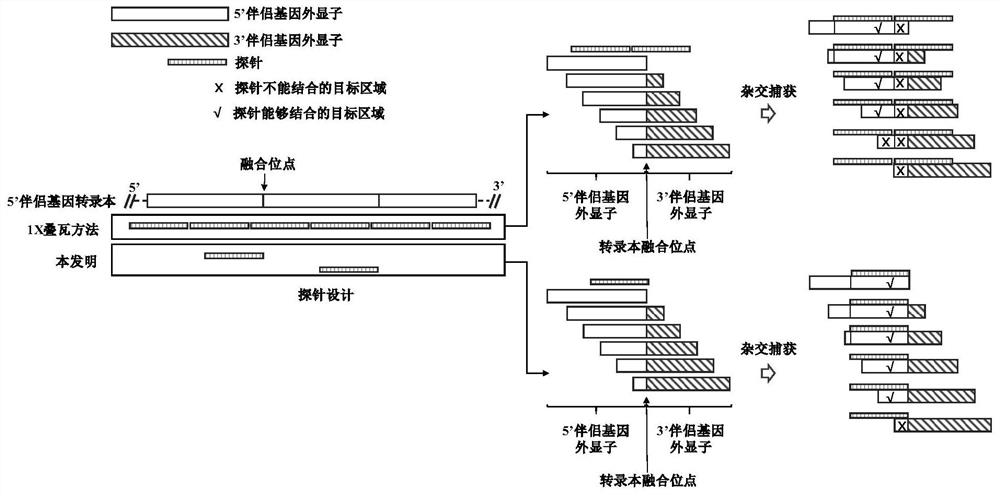
[0110] The traditional method and the method of the present invention were used to design probes for the genes related to 90 tumor gene fusion mutations, and part of the gene information is shown in Table 6 below; taking ALK as an example, Figure 4 The difference between the traditional method and the method of the present invention for probe design for ALK is shown, and the two use 39 and 29 120nt probes respectively, covering the interval of 4614bp and 3235bp; for the above 90 genes, the traditional method and the method of the present invention respectively 2075 and 1405 120nt probes were used; among them, the specific transcripts and probe information of ALK, RET and ROS1 genes are shown below.
[0111] Table 6 Traditional probe designs
[0112]
[0113]
[0114]
[0115]
[0116]
[0117] Table 7 Probe design of the present invention
[0118]
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com