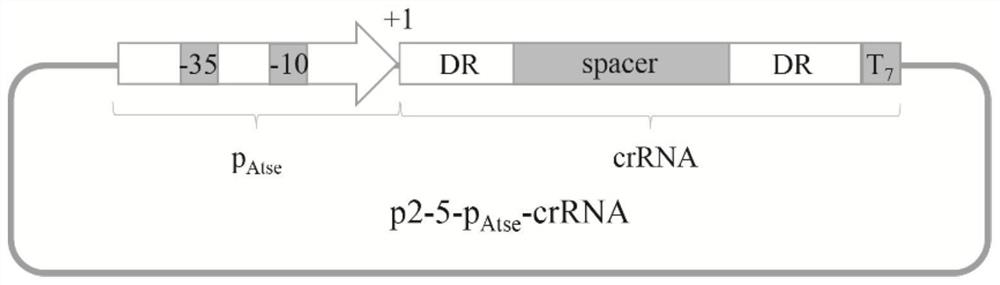
A method for gene suppression in Gluconobacter oxidans
A technology of gluconic acid bacteria and glucose oxidation, which is applied in the field of genetic engineering and bioengineering, can solve the problems of resistance integration constraints, many false positives of pentafluorouracil, and increased workload, so as to inhibit expression, improve gene editing efficiency, and effectively express Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
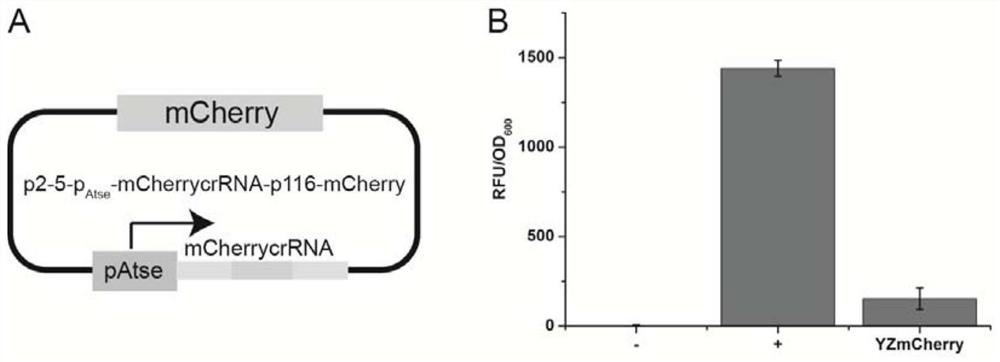
[0035] Embodiment 1: crRNA system inhibits the expression of mCherry in Gluconobacter oxidans
[0036] Designed for vector p2-5-p 116 - mCherry linearized primer pair F1 and R1,
[0037] F1:
[0038] atcctctggatgcaacgatcctgaagc ctcgtgtgttccccgcacacgcggggatgaaccgttttttttcgtcagcgggtgttggc (the underlined part is the homology arm sequence, the same below),
[0039] R1: tcaatattgagcccacggaaaactttcccacgccctgacgggcttg .
[0040] As laboratory preserved p2-5-p 116-mCherry plasmid (SEQ ID NO.2) was used as a template, and the primers were used for PCR linear amplification, and 2×Phanta Max Master Mix (Vazyme Company) was selected for pre-denaturation at 95°C for 3 minutes; amplification stage 30 cycles, according to 95°C, 15s, 56°C, 15s, 72°C, 4min; final extension at 72°C, 5min.
[0041] Designed to amplify p Atse - Primer pair F2 and R2 for mCherrycrRNA sequence
[0042] F2:
[0043] tgggaaagttttccgtgggc,
[0044] R2:
[0045] gcttcaggatcgttgcatccagaggat cggttcatcccc...
Embodiment 2
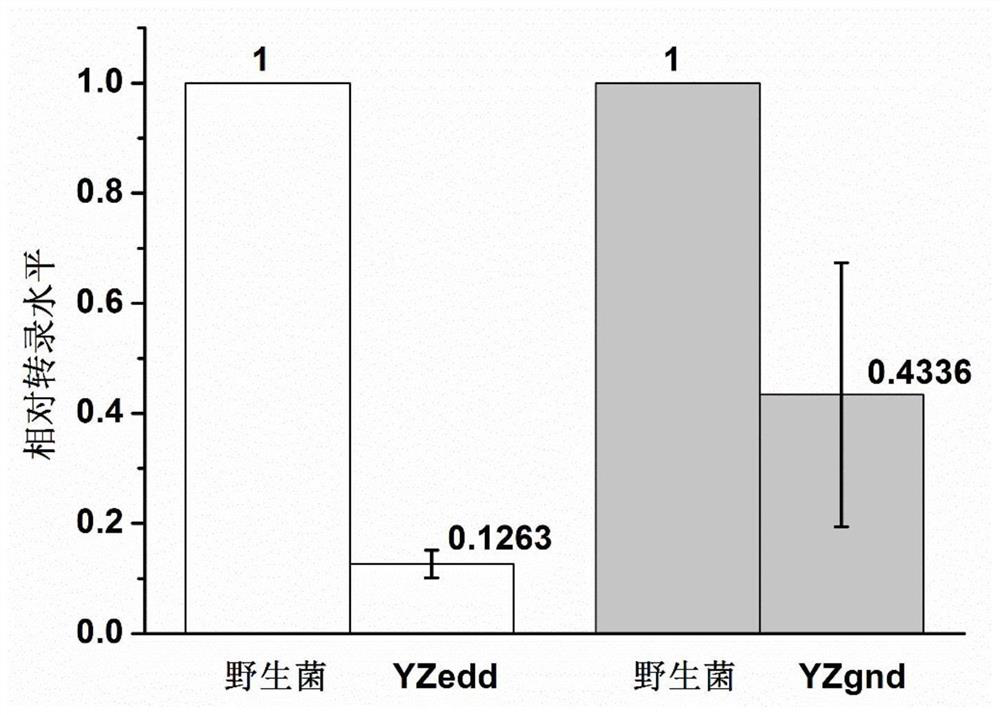
[0049] Embodiment 2: Use crRNA system to suppress the expression of edd in Gluconobacter oxydans
[0050] The gene edd (nucleotide sequence shown in SEQ ID NO.3) on the genome of Gluconobacter oxydans WSH-003 is weakened.
[0051] Primer F3 designed for linearization of vector p2-5,
[0052] F3:
[0053] gggcgtctcgcgtccgcgtctggcctgcggaaag tgttccccgcacacgcggggatgaaccgttttttttcctcttcgctattacgccag.
[0054] Utilize F3 and R1, use the p2-5 plasmid (SEQ ID NO.4) preserved in the laboratory as a template, use the primers to perform PCR linear amplification, select 2×Phanta Max Master Mix (Vazyme Company) to carry out, the condition Pre-denaturation at 95°C, 3min; 30 cycles of amplification, 95°C, 15s, 56°C, 15s, 72°C, 3min; final extension at 72°C, 5min.
[0055] Designed to amplify p Atse -crRNA edd sequence of primer R3,
[0056] R3:
[0057] tccgcaggccagacgcggacgcgagacgccc ggttcatccccgcgtgtgcggggaacacaaaccaatgaaaaataaagattattccgttc.
[0058] Using primers F2 and R3, a...
Embodiment 3
[0063] Embodiment 3: utilize crRNA system to suppress the expression of gnd in Gluconobacter oxydans
[0064] The gene gnd (nucleotide sequence shown in SEQ ID NO.5) on the genome of Gluconobacter oxydans WSH-003 is weakened.
[0065] Primer F4 designed for linearization of vector p2-5,
[0066] F4: gtgggcgcgccacgggggcagacacatt cgaggtgttccccgcacacgcggggatgaaccgttttttttcctcttcgctattacgcc.
[0067] Use F4 and R1, use the p2-5 plasmid stored in the laboratory as a template, and use the primers to perform PCR linear amplification on it. Select 2×Phanta Max Master Mix (Vazyme Company) for pre-denaturation at 95°C for 3 minutes. ; Amplification stage 30 cycles, according to 95 ℃, 15s, 56 ℃, 15s, 72 ℃, 3min; final extension 72 ℃, 5min.
[0068] Designed to amplify p Atse -crRNA gnd sequence of primer R4,
[0069] R4:
[0070] aatgtgtctgcccccgtggcgcgcccac cggttcatccccgcgtgtgcggggaacacaaaccaatgaaaaataaagattattccgttcg.
[0071] Using F2 and R4, and using the Gluconobacter oxi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com