Recombinant coccidiosis vector with MIC3 gene knocked out and detection method of recombinant coccidiosis vector
A detection method and technology of coccidia, which is applied in the field of genetic engineering, can solve the problems such as the unclear mechanism of MIC3 protein and coccidia infection, and achieve the effects of heritability and stability, length reduction and efficiency improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
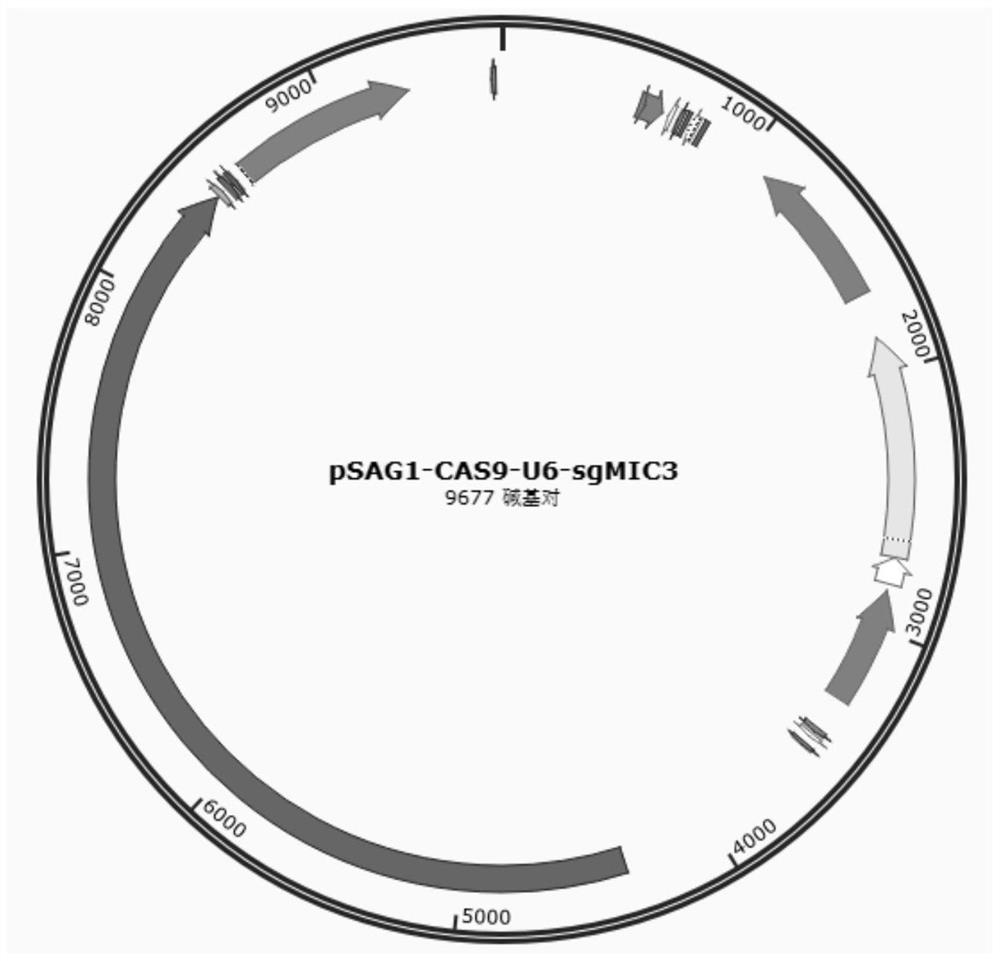
[0074] This embodiment provides a coccidia MIC3 gene editing system, the coccidia MIC3 gene editing system includes sgRNA targeting the coccidia MIC3 gene, Cas9 and a homologous recombination plasmid;
[0075] The sgRNA targeting the coccidian MIC3 gene is the nucleotide sequence shown in SEQ ID NO.1;
[0076] The sgRNA targeting the coccidian MIC3 gene is connected to the Cas9 in the same gene editing plasmid;
[0077] The homologous recombination plasmid includes a 5' homology arm, a SAG13 promoter, an EGFP gene and a 3' homology arm connected in sequence.
[0078] SEQ ID NO. 1: tacgaccccgcctactcctacgg.
[0079] The gene editing plasmid is prepared by the following method:
[0080] (1) PCR amplification of the coding sequence of the sgRNA and Cas9 targeting the coccidian MIC3 gene:
[0081] Using SEQ ID NO.3 and SEQ ID NO.4 as primers, and using the pSAG1::Cas9-U6::sgUPRT plasmid as a template, perform a PCR reaction to obtain a PCR product;
[0082] The reaction system ...
Embodiment 2
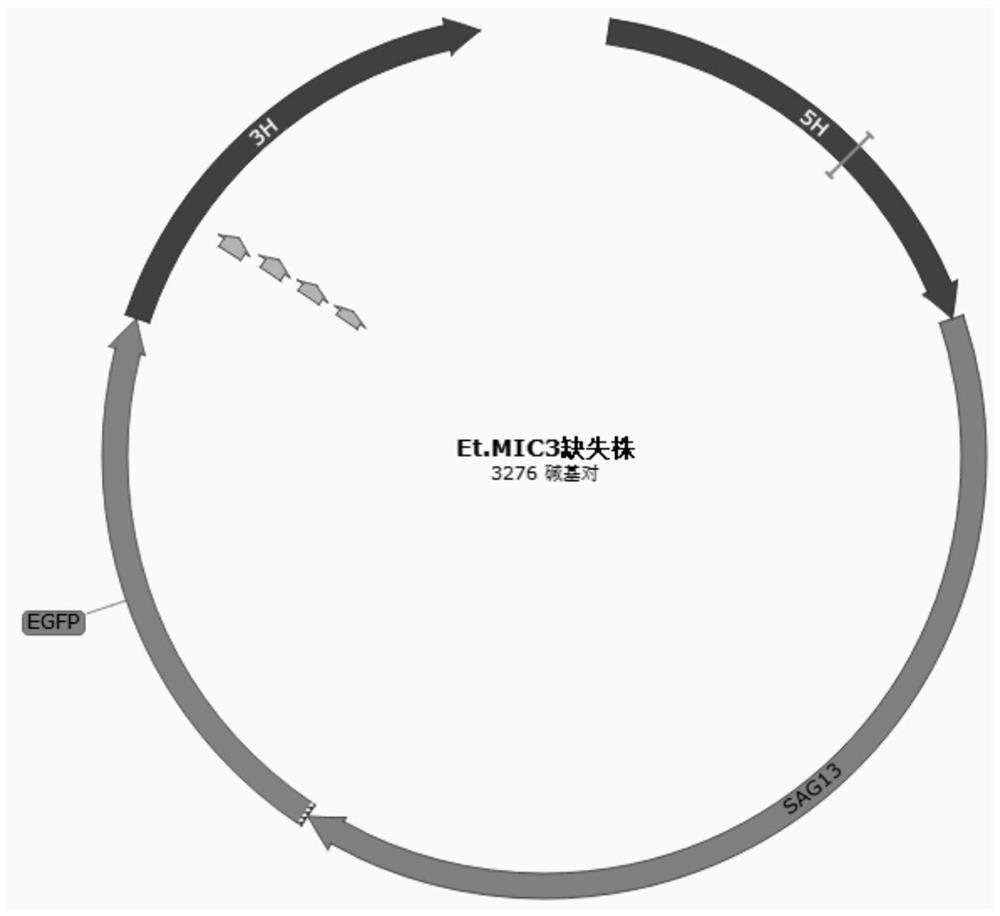
[0146] This embodiment provides a recombinant coccidia vector, which is a coccidia vector in which the EGFP gene is inserted into the coccidia MIC3 gene after being edited by the coccidia MIC3 gene editing system described in Example 1.
[0147] The schematic diagram of the construction principle of the recombinant coccidian vector is as follows: image 3 As mentioned, the build method is as follows:
[0148] (1) Prepare Eimeria tenella sporozoites:
[0149] Put the sporulated oocyst liquid in a 50mL centrifuge tube, centrifuge at 4000rpm for 10min, discard the supernatant, add sterilized PBS to wash 3 times, centrifuge at 4000rpm for 10min, discard the supernatant, and keep the precipitate;
[0150] Add 1 times the volume of 30% sodium hypochlorite solution to the precipitate, resuspend and let stand for 10 minutes, centrifuge at 1500 rpm for 10 minutes, collect the supernatant, and check the precipitate under a microscope. If there are more sporulated oocysts, add 30% sodiu...
Embodiment 3
[0166] In this embodiment, the recombinant coccidial vector prepared in Example 2 is detected, and the steps are as follows:
[0167] The recombinant coccidian oocysts were collected for genome extraction, and the genome was used as a DNA template to design primers PCR1-F (located in the 5'UTR region of ΔEtMIC3, SEQ ID NO.17), PCR1-R (located downstream of the SAG13 promoter of ΔEtMIC3, SEQ ID NO.17) IDNO.18), PCR2-F (located upstream of EGFP of ΔEtMIC3, SEQ ID NO.19), PCR2-R (located in the 3'UTR region of ΔEtMIC3, SEQ ID NO.20), PCR3-F (located inside the EtMIC3 gene, SEQ ID NO.21), PCR3-R (located inside the EtMIC3 gene, SEQ ID NO.22) for PCR reactions, the amplification system and procedures are the same as those in the construction of gene editing plasmids.
[0168] SEQ ID NO. 17: ttggcgcggaagtctgaagat;
[0169] SEQ ID NO. 18: cctttgctttctgtgtttttccgc;
[0170] SEQ ID NO.19: atggtgagcaagggcgag;
[0171] SEQ ID NO. 20: acccgcagttcaactctgttca;
[0172] SEQ ID NO. 21: aa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com