The method and application of judging background introduced microbial sequence
A technology of microorganisms and pathogenic microorganisms, which is applied in the field of bioinformatics analysis, can solve the problems of batch false positives, false negatives, and the inability to judge the origin of microorganisms from the sample itself, and achieve the effect of correcting quantitative errors in comparison and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] A method for judging background microbial sequences, comprising the following steps:
[0058] 1. Determine the list of background microorganisms.
[0059] 1. Sequencing.
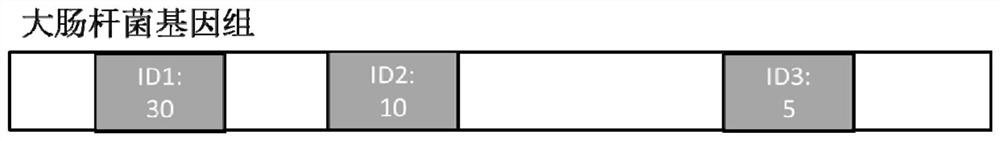
[0060] Obtain 1000 negative control plasma samples, extract nucleic acid and quantify the total amount of nucleic acid D, and then perform library sequencing.
[0061] mNGS off-machine data for microbial quantification: first use fastp software to remove adapters, low-quality sequences or sequences with a length less than 35bp; then use bwa software to compare to the human reference genome, and remove the sequences compared to the human reference gene; after that, The obtained non-human genome sequence (unHost Reads) was compared to the microbial database (including reference sequences of 18562 microorganisms) with bwa; finally, the quantitative value A of microbial abundance was obtained according to the number of aligned sequences.
[0062] 2. Determine the background microorganisms.
[0063] Acc...
Embodiment 2
[0092] 1464 clinical samples were collected as a verification set, among which 275 samples were detected with E. coli sequences (22 cases were clinically judged as E. coli positive samples), and the method of Example 1 was used to analyze and judge the sequence of microorganisms introduced into the background.
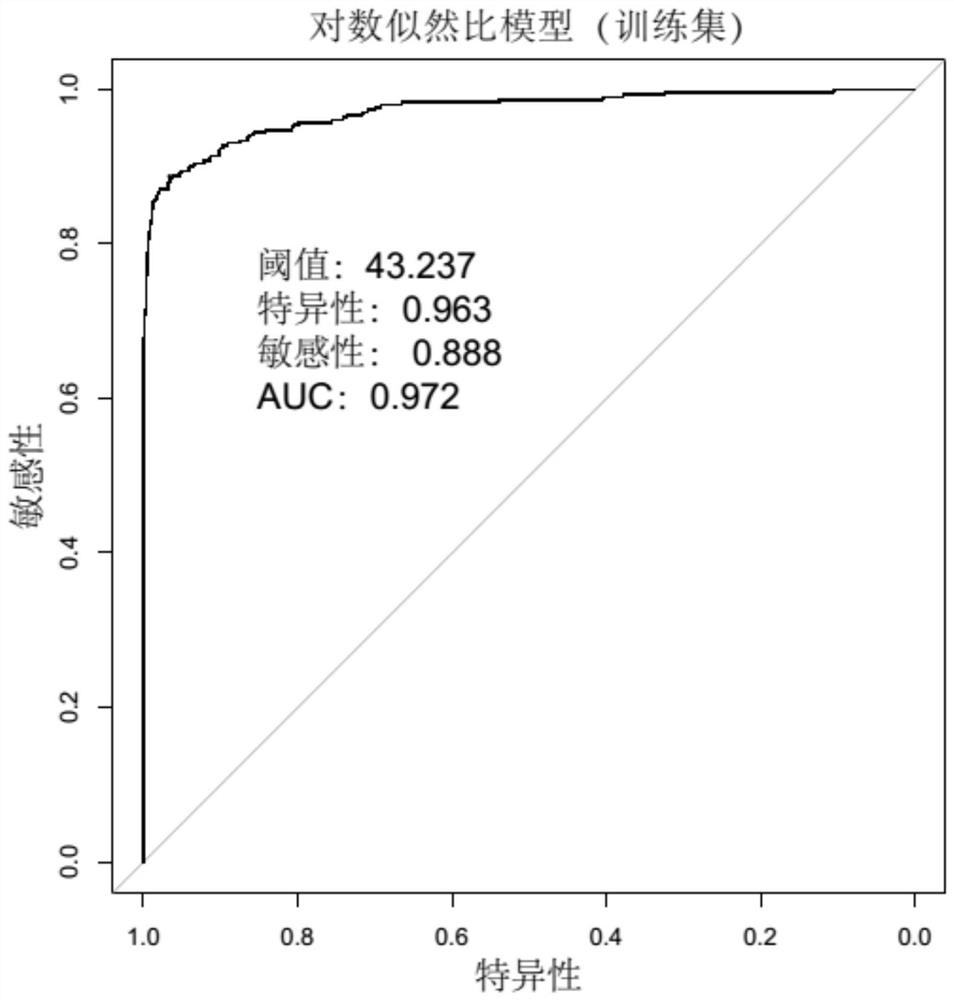
[0093] Sequence according to the method of Example 1, obtain the Escherichia coli gene sequence data, substitute it into the above judgment model, and judge with the log likelihood ratio LLR statistic in Example 1, the threshold is 40.684, and evaluate the classification effect of the model.
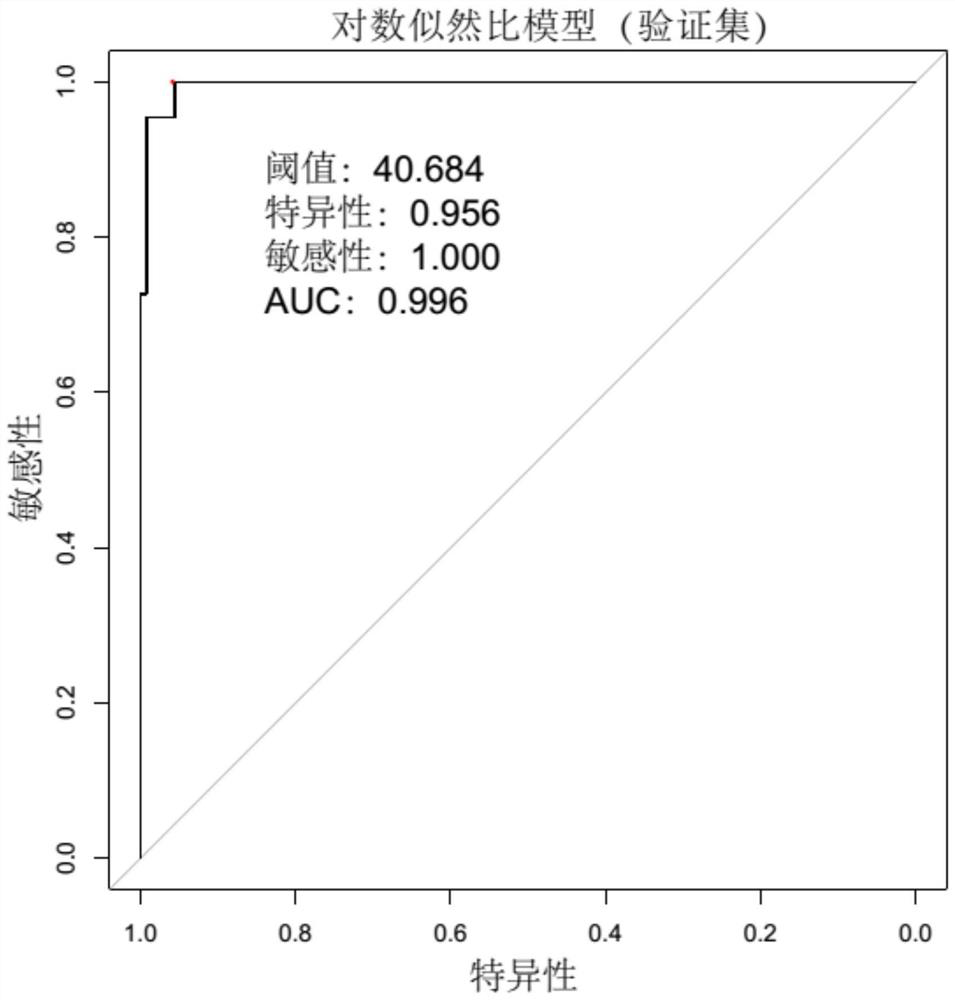
[0094] ROC curve such as image 3 As shown, the AUC value is 0.996, the specificity is 0.956, and the sensitivity is 1.000.
[0095] The above results show that the method of introducing microbial sequences into the judgment background of the present invention can achieve a better discrimination effect on samples from different source batches, and has the advantages of high judgm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com