Pathogen detection background microorganism judgment method and application
A microbiological and background technology, applied in the field of bioinformatics analysis, can solve the problems of batch false positives, fluctuations, high internal references, etc., and achieve the effect of wide range of functions and high application scalability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
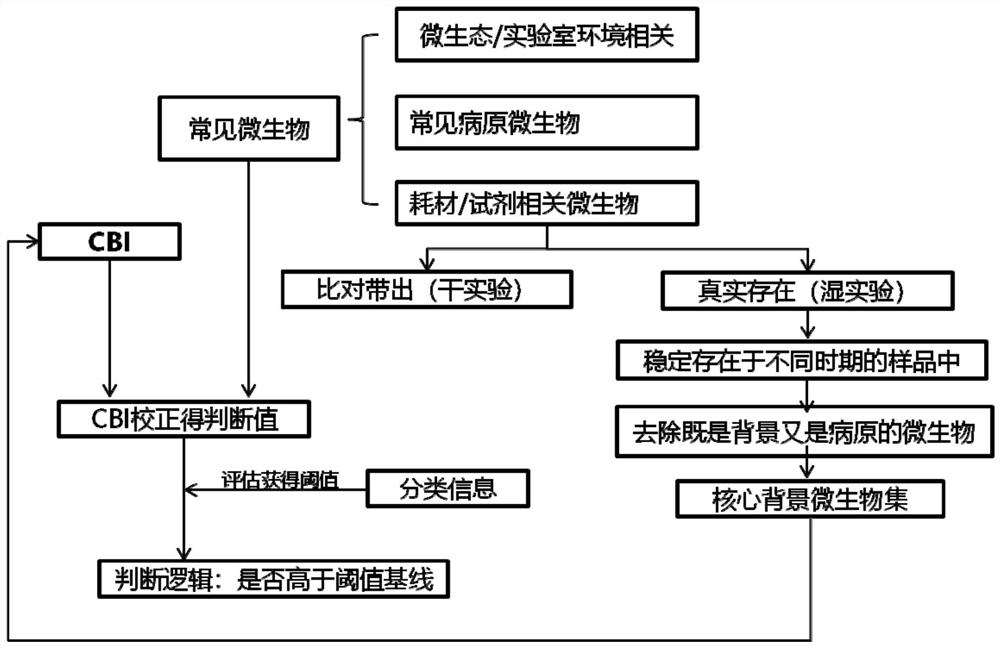
[0107] A method for determining pathogen detection background microorganisms, such as figure 1 shown, including the following steps:
[0108] 1. Determine the list of core background microorganisms.
[0109] 1.1 Sample analysis.
[0110] Take about 20,000 mNGS DNA samples sequenced in our company's Guangzhou laboratory. According to the time sequence of the samples, every 2,000 samples are used as a data set (each data set contains various types of samples at the same time, such as Alveolar lavage fluid, sputum, cerebrospinal fluid, throat swab, blood, tissue, etc.).
[0111] Firstly, obtain the microorganisms whose occurrence frequency is greater than 25% in the data set and appear in all the data sets, that is, the microorganisms that appear frequently and stably in the samples.
[0112] Then calculate the pearson correlation test and spearman correlation test between the nucleic acid extraction concentration or library concentration of these samples and the number of spe...
Embodiment 2
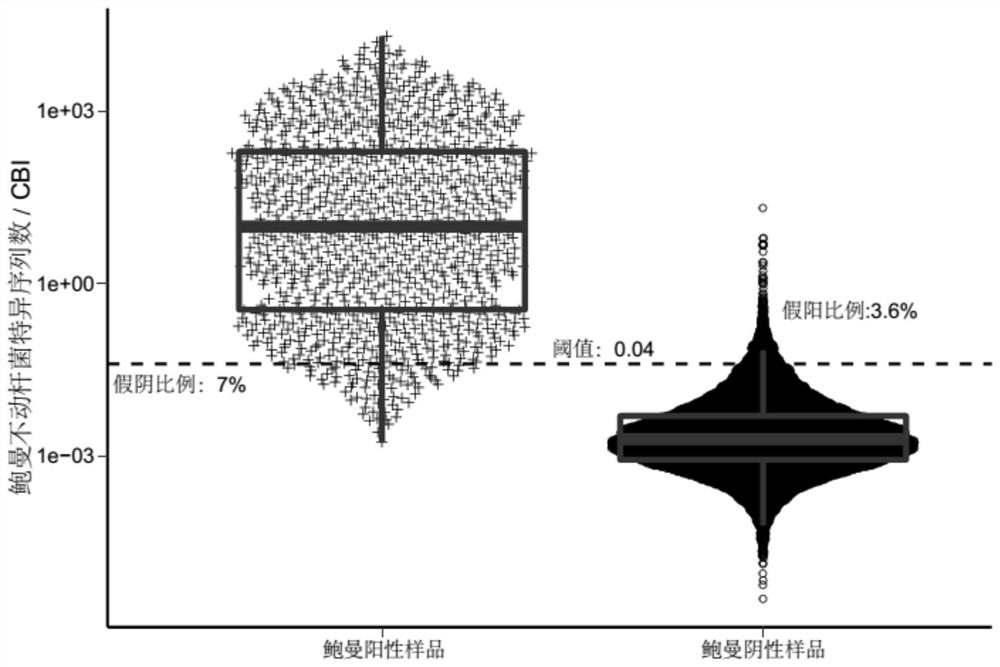
[0138] 639 clinical samples were collected as a verification set, and the method in Example 1 was used to analyze and judge whether the characteristic sequence in the sample was a background sequence.
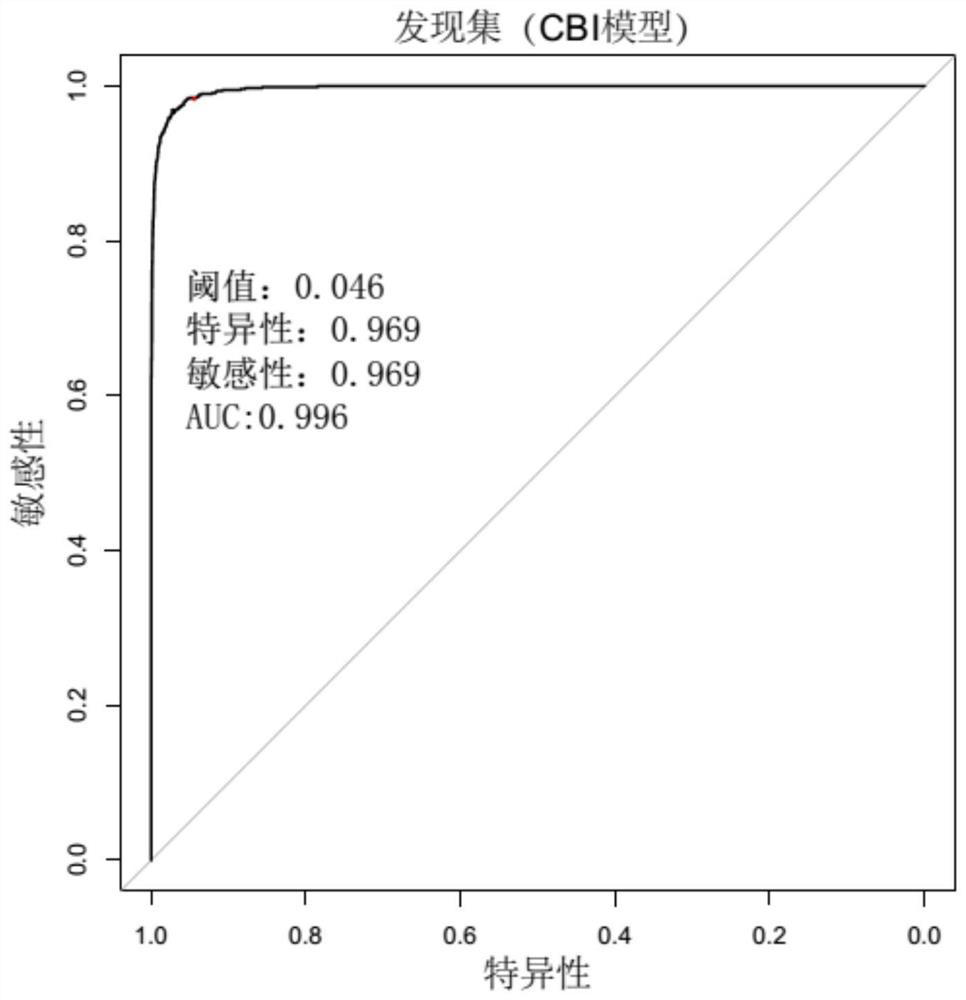
[0139] Sequence according to the method of Example 1, obtain wherein Acinetobacter baumannii gene sequence data, analyze according to the above-mentioned method, build a model, draw the ROC curve, the result is as follows Figure 4 As shown, judging by the threshold value of 0.041, the AUC value was 0.992, the specificity was 0.942, and the sensitivity was 0.983.
[0140] The above results show that the method for determining background microorganisms in pathogen detection of the present invention can achieve a better discrimination effect on samples from different source batches, and has the advantages of high determination stability and strong practicability.
[0141] It can be understood that the judgment method in the above embodiment is applicable to a wide range of sample...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com