Pathogen detection background microorganism judgment method and application
A microbial and background technology, applied in the field of bioinformatics analysis, can solve problems such as fluctuations, batch false positives, background pollution, etc., and achieve high application scalability and wide-ranging effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
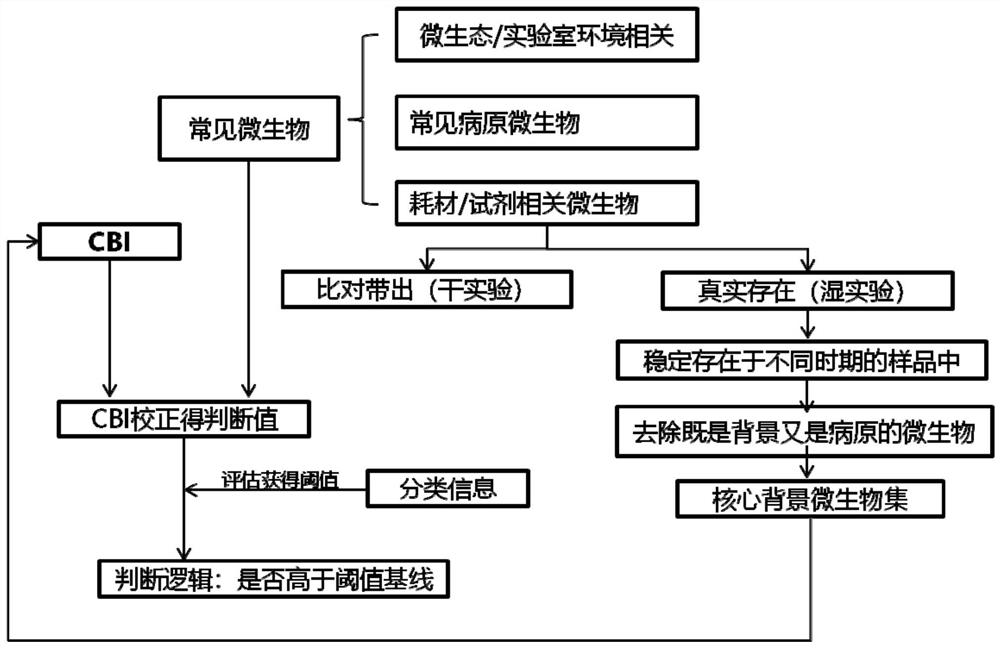
[0107] A method for determining pathogen detection background microorganisms, such as figure 1 shown, including the following steps:
[0108] 1. Determine the list of core background microorganisms.
[0109] 1.1 Sample analysis.
[0110] Take about 20,000 mNGS DNA samples sequenced in our company's Guangzhou laboratory. According to the time sequence of the samples, every 2,000 samples are used as a data set (each data set contains various types of samples at the same time, such as Alveolar lavage fluid, sputum, cerebrospinal fluid, throat swab, blood, tissue, etc.).
[0111] Firstly, obtain the microorganisms whose occurrence frequency is greater than 25% in the data set and appear in all the data sets, that is, the microorganisms that appear frequently and stably in the samples.
[0112] Then calculate the pearson correlation test and spearman correlation test between the nucleic acid extraction concentration or library concentration of these samples and the number of spe...
Embodiment 2
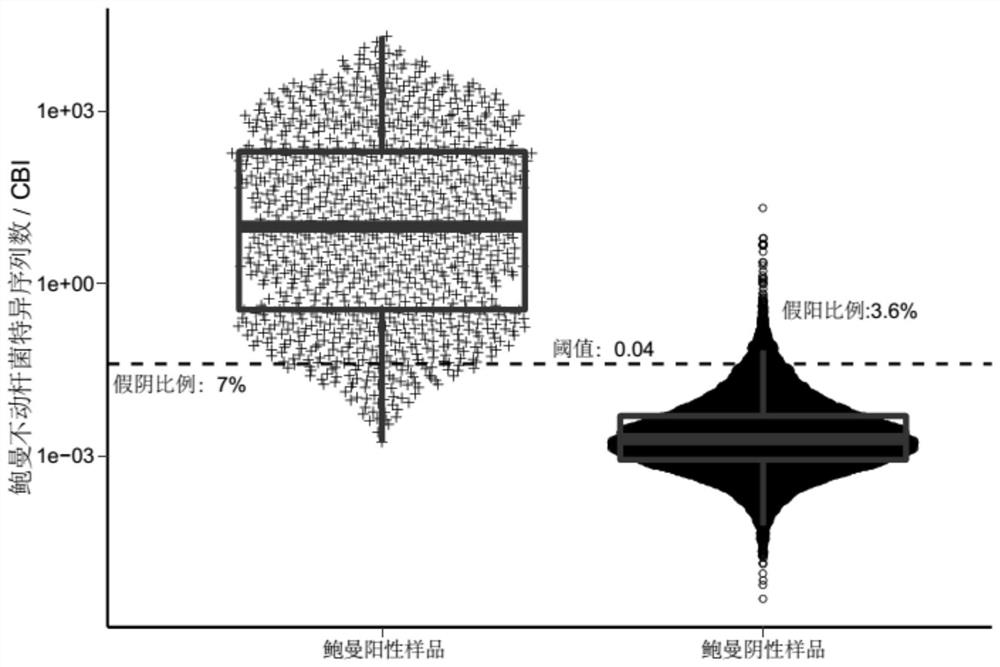
[0138] 639 clinical samples were collected as a verification set, and the method in Example 1 was used to analyze and judge whether the characteristic sequence in the sample was a background sequence.
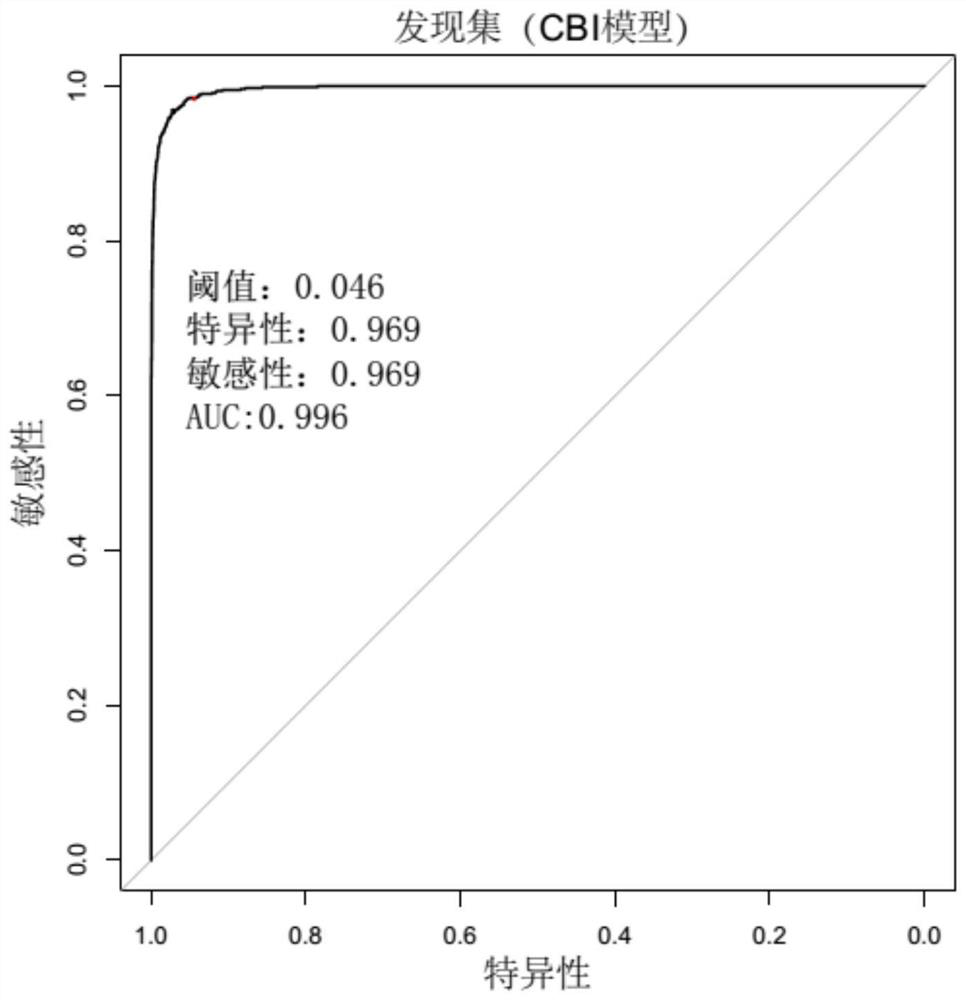
[0139] Sequence according to the method of Example 1, obtain wherein Acinetobacter baumannii gene sequence data, analyze according to the above-mentioned method, build a model, draw the ROC curve, the result is as follows Figure 4 As shown, judging by the threshold value of 0.041, the AUC value was 0.992, the specificity was 0.942, and the sensitivity was 0.983.
[0140] The above results show that the method for determining background microorganisms in pathogen detection of the present invention can achieve a better discrimination effect on samples from different source batches, and has the advantages of high determination stability and strong practicability.
[0141] It can be understood that the judgment method in the above embodiment is applicable to a wide range of sample...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com