Nucleic acid detection method based on liquid chip technology
A detection method and liquid chip technology, applied in the field of molecular biology, can solve the problems of time-consuming and high R&D costs, and achieve the effects of reducing the burden of instruments, increasing safety, and saving R&D time and R&D costs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
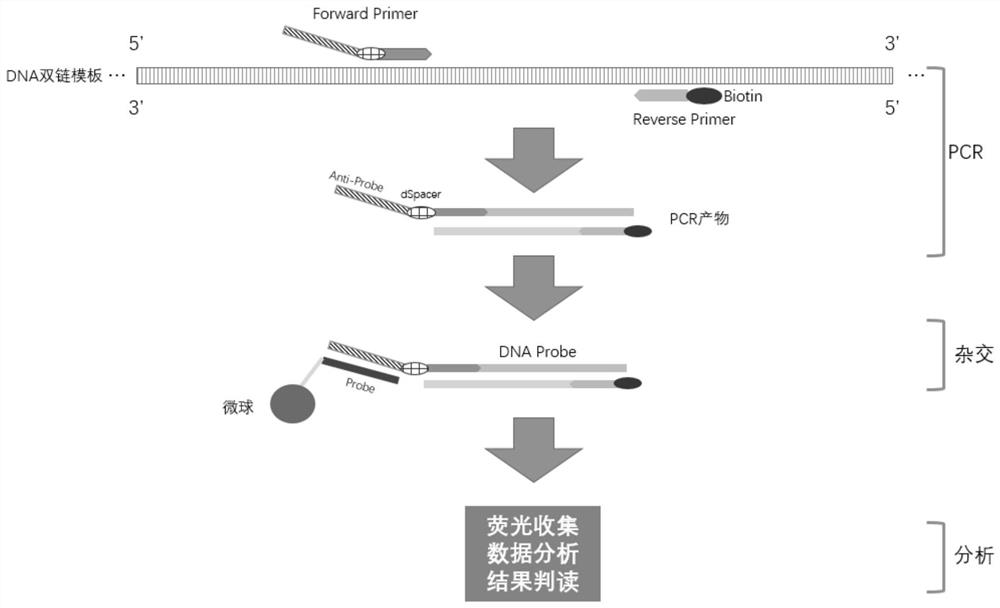
[0025] This embodiment provides a nucleic acid detection method based on liquid chip technology. This method combines multiplex PCR technology, constant temperature hybridization based on liquid chip technology, detection technology and specific sequence, and the principle of coding microspheres. The typing detection kit for different mutant strains of coronavirus can realize the double-stranded DNA product obtained after PCR after the reverse transcription of the nucleic acid RNA template of the new coronavirus (Bio-Rad reverse transcription reagent is used, see Table 2 for the configuration of the reverse transcription system), And realize the hybridization and signal detection of the probe system under the condition of constant temperature. The detection process includes steps:
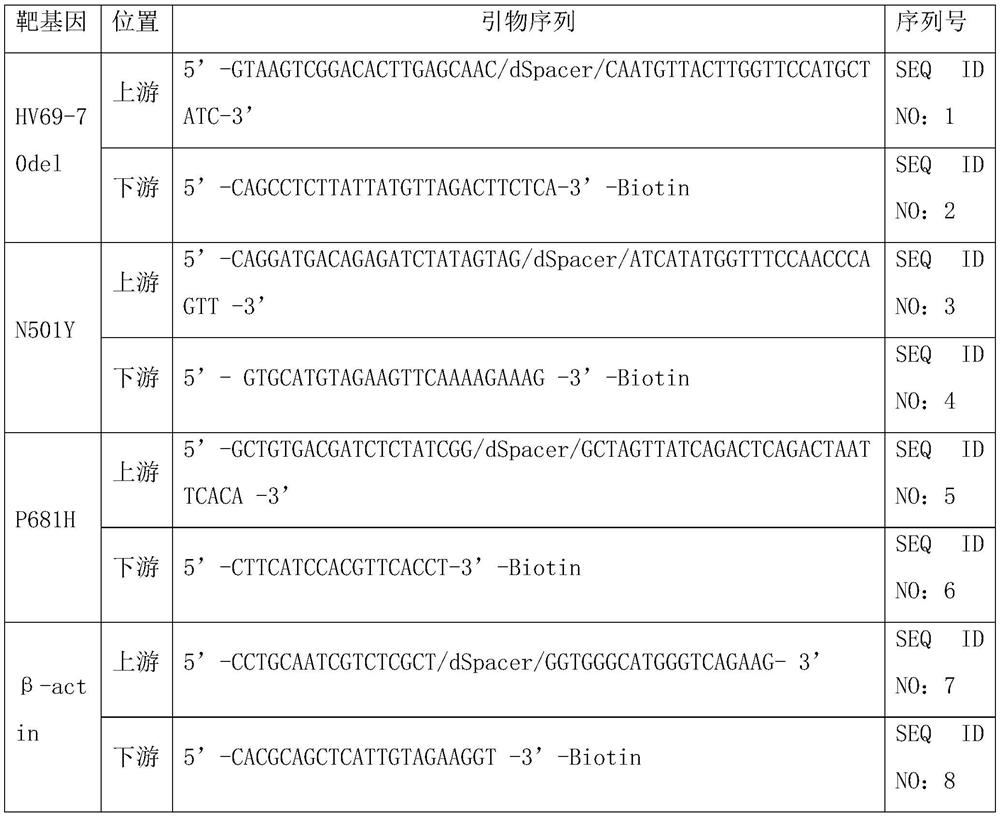
[0026] 1) Amplify the pseudoviruses of the mutation sites of the new coronavirus HV69-70del, N501Y and P681H with a labeled primer and a common primer connected to the specific coding Anti-Probe se...
Embodiment 2
[0053] This embodiment provides a nucleic acid detection method based on liquid chip technology. This method combines multiplex PCR technology, constant temperature hybridization based on liquid chip technology, detection technology and specific sequence, and the principle of coding microspheres. The HPV typing detection kit can realize the double-stranded DNA product obtained after the human papillomavirus DNA template is enriched by multiple PCR target sequences, and realize the hybridization and signal detection of the probe system under constant temperature conditions . The detection process includes steps:
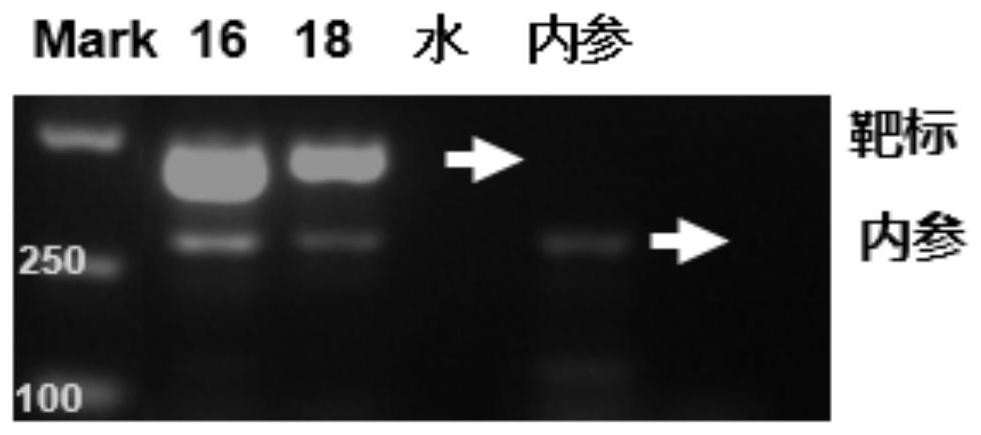
[0054] 1) Amplify HPV16, HPV18, and internal reference β-Globin templates with a labeled primer and a common primer connected to a specific coding Anti-Probe sequence (see Table 10 for the specific primer sequence) (the template is synthesized according to NCBI search DNA sequence), the PCR program is as shown in Table 12, and the PCR system preparation is as shown i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com