Pyridoxine phosphate synthase pdxj mutant and its application in the preparation of vitamin b6
A technology of pyridoxine phosphate and mutant, applied in the biological field, can solve problems such as limiting fermentation yield, and achieve the effect of improving the ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1: Construction of the original gene of pyridoxine phosphate synthase PdxJ
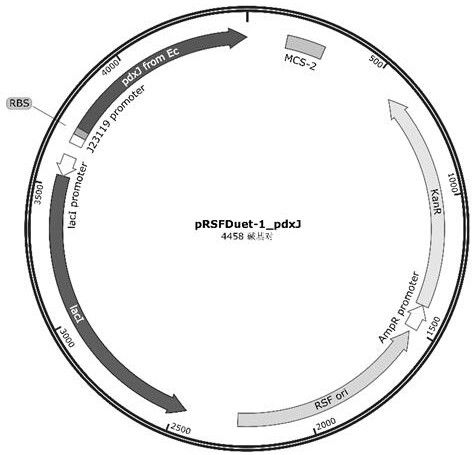
[0054] 1. Amplify the original gene of pyridoxine phosphate synthase PdxJ whose nucleotide sequence is as shown in SEQ ID NO: 2, the nucleotide sequence of which is shown in SEQ ID NO: 2, and the primers used For liulx-1 / liulx-2, the wild-type pRSFDuet-1 plasmid was used as a template to amplify the backbone (primers liulx-3 / liulx-4), and the PdxJ gene was linked to the plasmid backbone by Gibson assembly, transformed into DH5α E. coli, and coated On the LB plate (containing 50 μg / mL kanamycin), screen the positive clones and verify that the bands (primers DuetUP1 / Duetdown1) are correct and sequenced to confirm the correct recombinant plasmid pRSFDuet-1_pdxJ original gene vector (for the map, see figure 1 ), and the plasmid was extracted for use.
[0055] Table 1 Primers constructed by PdxJ original gene vector
[0056]
Embodiment 2
[0057] Example 2: Design of pyridoxine phosphate synthase PdxJ mutation site
[0058] First, the homology alignment analysis was performed between the pyridoxine phosphate synthase PdxJ derived from Escherichia coli and the amino acid sequence of pyridoxine phosphate synthase reported in the Genbank database. 1M5W) analysis and understanding of the catalytic mechanism, simulate the docking mode between the substrate and the enzyme, and select a binding mode similar to the catalytic binding mechanism according to the docking results. For the designed residues, enzyme design was performed using Rosetta to design the amino acid sequence of the mutant.
Embodiment 3
[0059] Example 3: Construction of pyridoxine phosphate synthase PdxJ mutants
[0060] 1. A site-directed mutagenesis site was introduced by a one-step PCR method, and the recombinant plasmid pRSFDuet-1_pdxJ obtained in Example 1 was originally used as a template for single-site-directed mutagenesis. Three point mutants are plasmid templates. The basic process is to first design mutation primers (see Table 2 for primers), introduce mutation sites into the primers, perform overlap PCR, and then use Dpnl enzyme to identify methylation sites and digest them with the template. PCR treated with Dpnl enzyme The product was transformed, and finally the bacteria were selected for sequencing verification, and the correct plasmid was extracted for use.
[0061] Primers used in table 2 embodiment 3
[0062]
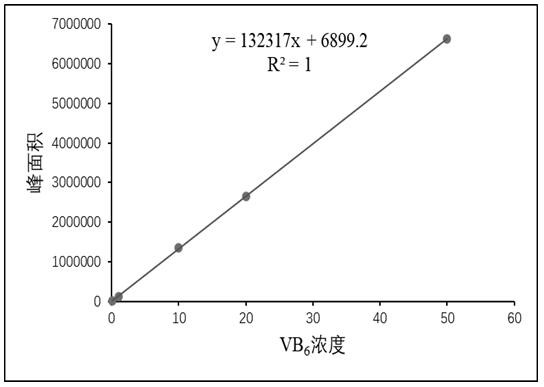
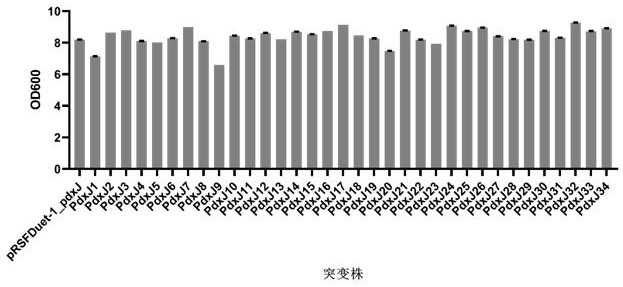
[0063] 2. A total of 34 PdxJ mutants were obtained and named as PdxJ1-pdxJ34. The amino acid differences of PdxJ1-pdxJ34 compared with the pRSFDuet-1_pdxJ original gene are sho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com