Method for carrying out polynucleotide variation identification and annotation on multiple species
A polynucleotide and multi-species technology, applied in the field of polynucleotide variation identification and annotation, can solve problems such as MNV annotation that cannot be larger than double points, and achieve the effect of reducing time and avoiding errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0055] In order to facilitate the understanding and implementation of the present invention by those of ordinary skill in the art, the present invention will be further described in detail below with reference to the embodiments. It should be understood that the embodiments described herein are only used to illustrate and explain the present invention, but not to limit the present invention.
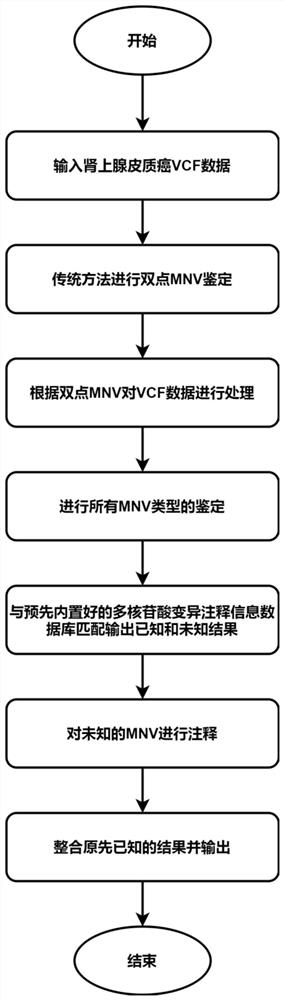
[0056] In this embodiment, a method for identifying and annotating polynucleotide variants for multiple species is to identify and annotate MNVs from species-oriented standard variant call format data (the variant call format, VCF). Specifically, as figure 1 shown, follow the steps below:
[0057] Step 1. Use the directional data in standard variant format of adrenocortical carcinoma (hereinafter referred to as data set VCF). First, use the traditional polynucleotide variant identification tool (identify_mnv, https: / / github.com / macarthur-lab / gnomad_mnv) to obtain all two-point MNVs as t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com