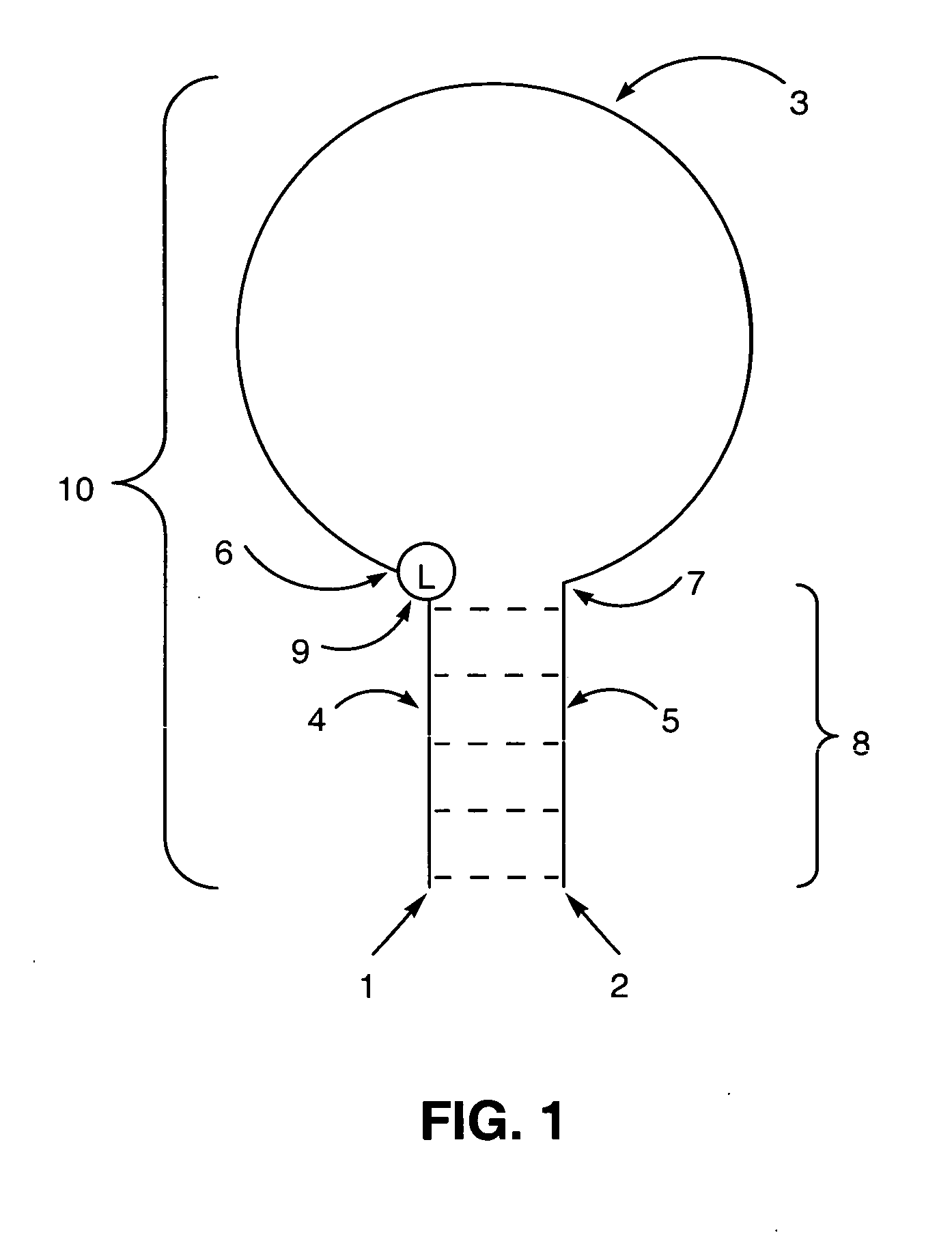
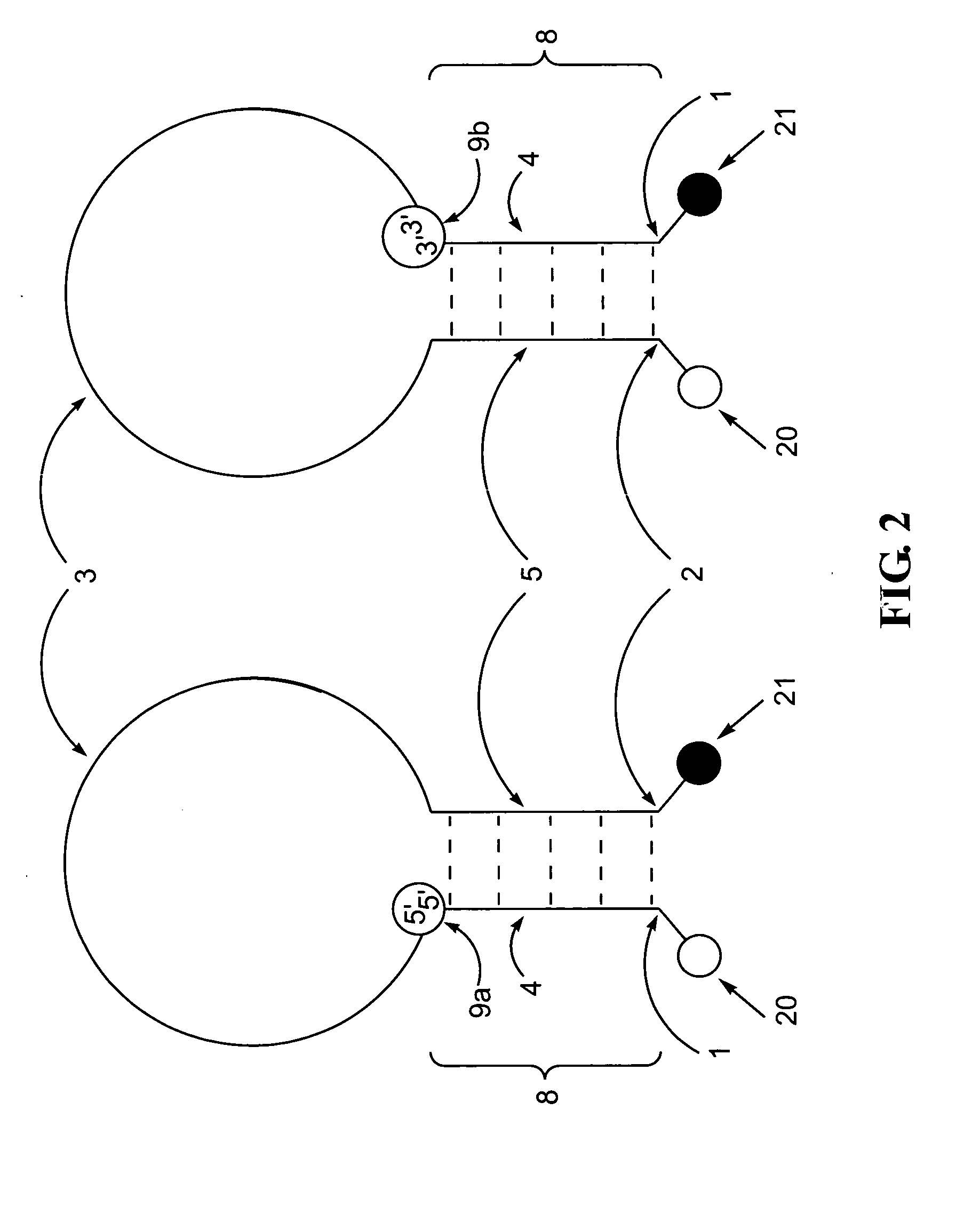
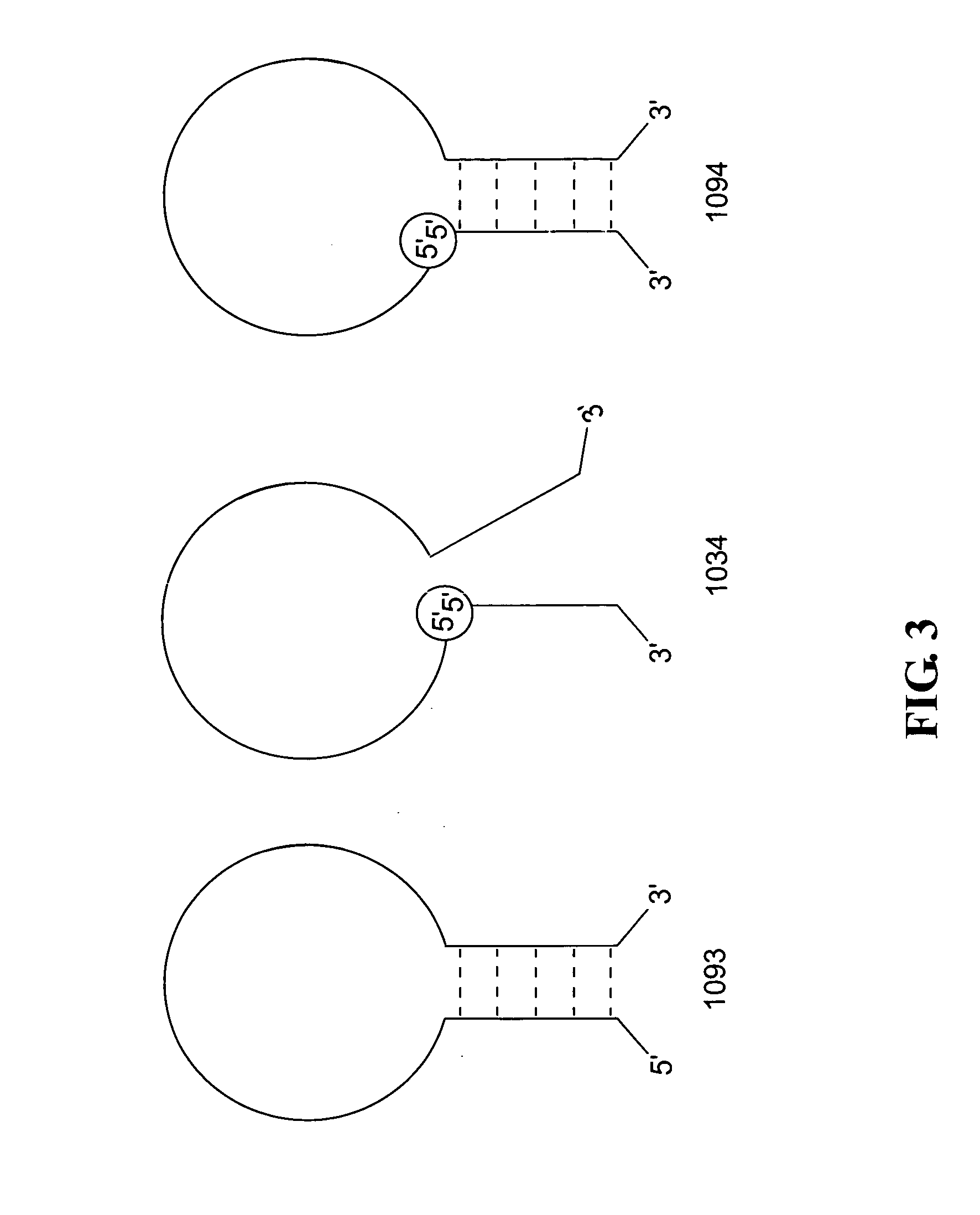
Polynucleotide detection method employing self-reporting dual inversion probes
a detection method and probe technology, applied in the field of nucleic acid detection, can solve the problem that the design of the molecular beacon probe is naturally rendered more complicated than the one befor
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Quantifying Interactions Between Polynucleotide Targets and Probes
[0098] The 1034, 1094 and 1093 probes were independently synthesized by solid-phase phosphite triester chemistry using DABCYL-linked controlled pore glass and 5′ fluorescein-labeled phosphoramidite on a Perkin-Elmer (Foster City, Calif.) EXPEDITE model 8909 automated synthesizer. The inversion linkages in the 1034 and 1094 probes, like all of the inversion linkages described herein, were created using a combination of 5′-β-cyanoethyl and 3′-β-cyanoethyl phosphoramidites that were purchased from Glen Research Corporation (Sterling, Va.), Proligo (Boulder, Colo.) or Pierce Biotechnology (Rockford, Ill.). All of these probes were constructed using 2′-OMe nucleotide analogs in their target-complementary loop regions, and standard deoxyribonucleotides in their stem regions. Following synthesis, the probes were deprotected and cleaved from the solid support matrix and then purified using polyacrylamide gel electrophoresis ...
example 2
Target Polynucleotide Binding Triggers Signaling by Parallel-Stem Hybridization Probes
[0107] Individual samples containing the 1093 molecular beacon or the 1094 parallel-stem hybridization probe and either of the two polynucleotide targets described in Example 1 were hybridized and monitored for fluorescence emission. In these procedures the concentrations of the probes were held constant at 0.3 μM, while the concentration of the target varied from 0-3 μM. Parallel procedures were carried out using the 1059 and 1061 targets. In all instances the mixtures were heated to 60° C. for 30 minutes, cooled to 42° C. for 30 minutes, and finally cooled to room temperature (about 23° C.) for 30 minutes before reading fluorescence at room temperature. These thermal step procedures promoted interaction between the probe and the target polynucleotide in the samples that included a target polynucleotide. Fluorescence measurements were carried out using a FLUOROSKAN ASCENT fluorometer (Labsystems,...
example 3
Parallel-Stem Hybridization Probes and Molecular Beacons Exhibit Different Functional Characteristics in Hybridization Assays
[0113] Samples containing the 1093 molecular beacon, the 1094 parallel-stem hybridization probe, or a combination of these two probes were hybridized at one of several probe concentrations with 0-0.3 μM of the 1059 target polynucleotide essentially as described above. Samples containing the 1093 molecular beacon had probe concentrations of either 0.1 μM, 0.15 μM, 0.2 μM, 0.25 μM or 0.3 μM. Samples containing the 1094 parallel-stem hybridization probe had probe concentrations of 0.05 μM, 0.1 μM, 0.15 μM or 0.2 μM. Samples containing both the molecular beacon and the parallel-stem hybridization probe all had total probe concentrations of 0.3 μM, with different proportions of this total being due to each of the two probes. In all cases background fluorescence measurements from buffer controls were subtracted from the fluorescence signals measured for each sample...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting temperatures | aaaaa | aaaaa |
| melting temperatures | aaaaa | aaaaa |
| Tm | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com