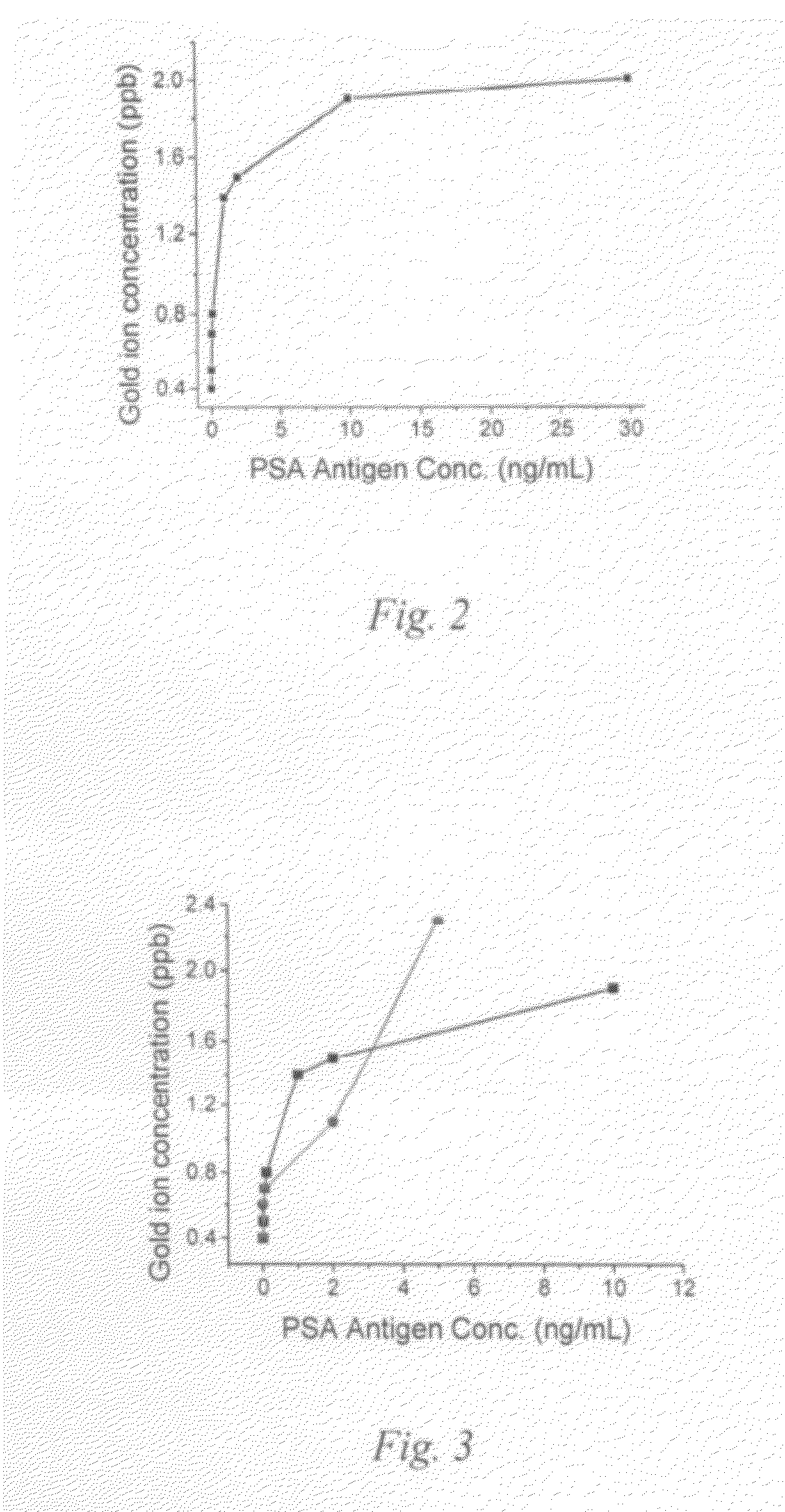Detection of biotargets using bioreceptor functionalized nanoparticles
a biomolecule and functional technology, applied in the field of biotarget detection systems and methods, can solve the problems of limited amplification effect of bar-code methods, inability to fully understand the effect of raman enhancement, and use of expensive biomolecules (dna) and rather sophisticated procedures and analytical instruments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0059]Currently, GFAAS and ICP-MS are the two most sensitive techniques for ultra trace metal analysis. The detection limit of GFAAS was examined on gold nanoparticles. A citrate stabilized nanoparticles with an average core diameter around 35 nm were decomposed by a solution of I2 / KI, where the I2 oxidizes gold atoms from the nanoparticles into gold ions. This solution was then diluted into four concentrations ranging from 10−11 to 10−15 M. This nanoparticle concentration range corresponds to a gold ion concentration ranging from 10−5 to 10−9 M. The analysis of 20 μL samples demonstrated that the lowest concentration of gold ions that can be detected by GFAAS is at the order of 10−8 M as analyzed using a Perkin Elmer AAalyst 600 instrument, resulting from a gold nanoparticle concentration of 10−14 M (10 s of femtomolar). Therefore, the minimal number of gold nanoparticles that can be detected by GFAAS is approximately 10,000.
[0060]Similar analysis was conducted on silver nanopartic...
example 2
[0061]PSA is a 32 kDa single chain glycoprotein serine protease with a chymotrypsin like specificity produced by the secretory epithelium of the prostate gland. PSA that is normally secreted into the seminal fluid plays a functional role in the cleavage of the seminal vesicle proteins and the liquefaction of the seminal coagulum. Only low levels of PSA are normally present in the blood stream, and increasing serum concentrations indicate prostatic pathology, including benign prostatic hyperplasia and cancer of the prostate. Determination of PSA is now widely used for detection and management of patients with prostatic cancer and considered as the superior serological marker for cancer of the prostate. The atomic amplification assay approach was used to examine the detection of PSA and compared the results with standard ELISA assay.
[0062]A commercial ELISA kit for PSA assay along with a paired detector antibody was purchased from AnoGen, Inc. We conducted PSA assay using the ELISA ki...
example 3
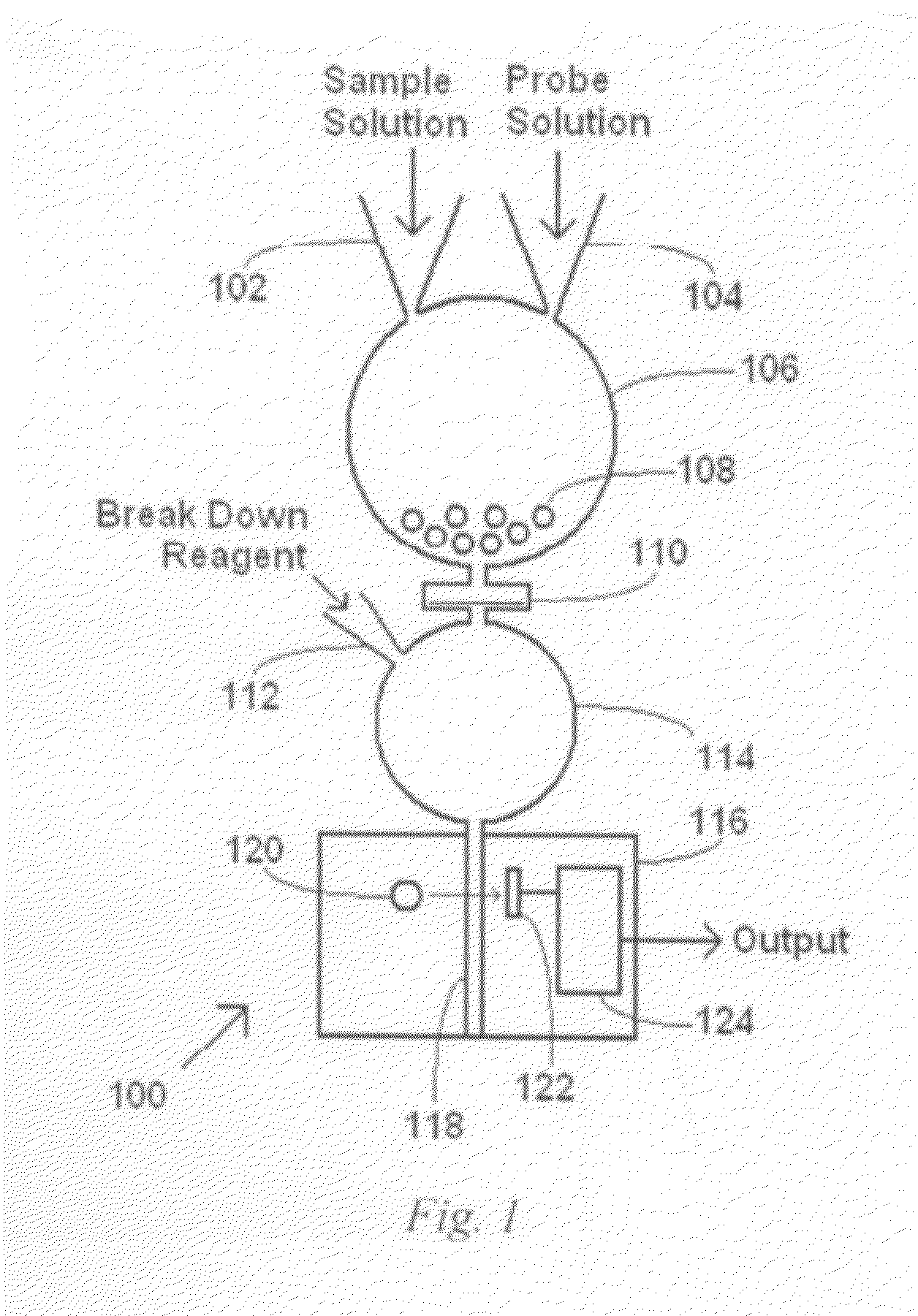
[0066]A route to nanoparticle probes according to an embodiment of the invention is given in FIG. 4. Relatively monodispersed citrate-stabilized gold nanoparticles can be synthesized using the method of Turkevich et al. in aqueous solution with sizes ranging from 10-120 nm, by controlling the volume of the capping ligand, tri-sodium citrate. The synthesis of citrate-stabilized silver nanoparticles can be done using by using silver nitrate as in Lee et al. J. Phys. Chem. 86 (1982) 3391. After purification, citrate-stabilized gold or silver nanoparticles will be converted to thioctic acid-protected nanoparticles through the ligand place exchange reaction. Two thiol groups-terminated thioctic acid is used to provide structural stability to the nanoparticles. The carboxyl groups on the nanoparticles are then activated with EDC (N-(3-dimethylaminopropyl)-N′-ethyl-carbodiimide) and NHS (N-hydroxysuccinimide). Two types of oligo(ethylene glycol) amine ligands, one with two amino end groups...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentrations | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com