Rapid aneuploidy detection
a technology of aneuploidy and rapid detection, applied in the field of gene analysis, can solve problems such as life-long challenges to families and societies, and achieve the effect of sensitive and rapid assessment of genomic copy number
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Methods
Primer Selection
[0030]We began by searching a −6.8 Mb region of chromosome 21 containing the Down's Syndrome critical region (hg1917 coordinates 35,888,786 to 42,648,523) for sequence blocks of ˜150 by that were similar but not identical to those present on all chromosomes. Using 150 by sliding windows incremented by 50 by (135,186 sequences of 150 by in length), we queried sequences with the BLAST-Like Alignment Tool (BLAT) algorithm13 to identify such blocks. We also required that at least three of the blocks were present on chromosome 21 in addition to the one within the ˜6.8 Mb region described above.
[0031]Out of the 135,186 queried blocks, we found only 56 that met the following criteria:[0032]contained at least 11 variant bases from the query sequence, to aid in distinguishing amplified loci;[0033]contained no more than 30 variant bases from the query sequence, to increase the chance of uniform amplification; and[0034]spanned no more than a total of 180 bases, to be com...
example 2
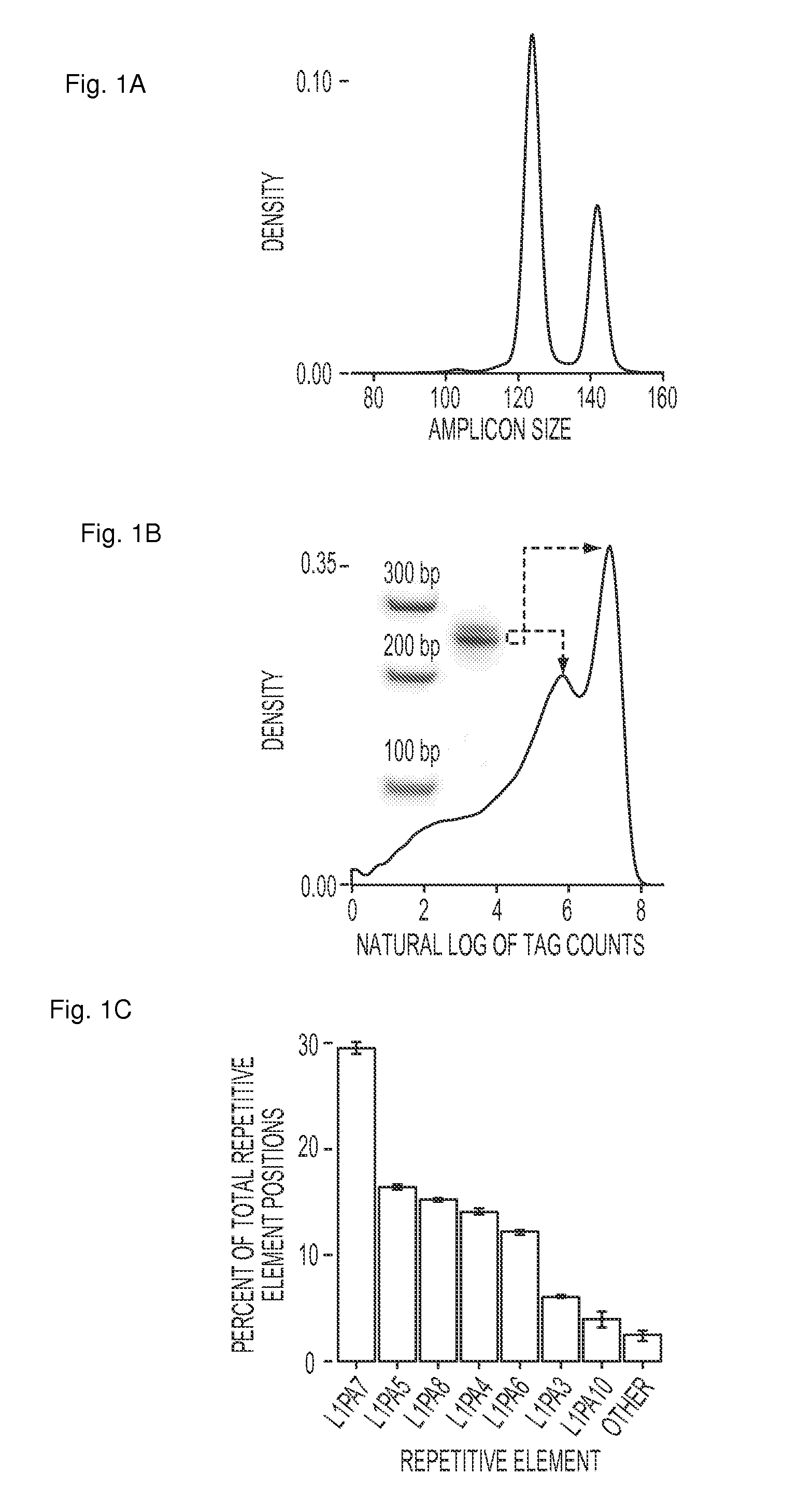
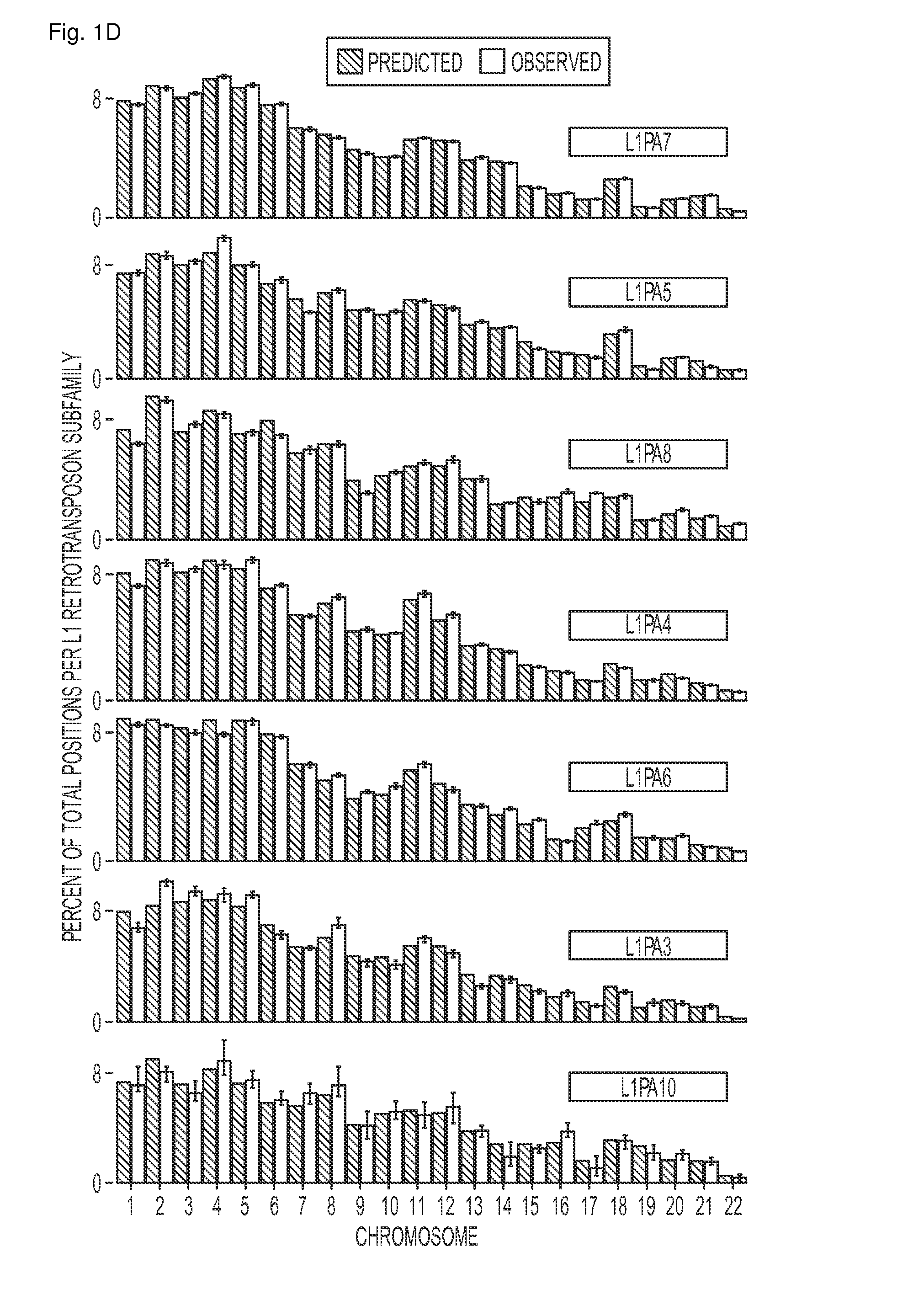
[0057]As an initial test of the performance of FAST-SeqS, we examined the representation of each autosome in the plasma DNA of seven normal females, including one biologic replicate (Supplementary Table 1). Using only 37 cycles of sequencing in one-quarter of a lane on an lumina HiSeq 2000 instrument, we recovered an average of 31,547,988 high quality tags per individual (range: 27,179,424 to 36,0418,017 tags; Supplementary Table 2). An average of 35% of these tags (range: 31 to 37%) could be uniquely mapped to one of an average of 23,681 unique chromosomal positions (range: 22,589 to 24,562 positions) when allowing up to one mismatch during alignment to hg1917 using Bowtie18. The theoretical in silico (FIG. 1a) and observed distribution of tag counts (FIG. 1b) both showed a bimodal distribution of sizes. Of the uniquely aligned tags, 99.1% aligned to positions predicted to be repetitive DNA by RepeatMasker (http: / / www.repeatmasker.org), 97.5% of which fell into just seven L1 retrot...
example 3
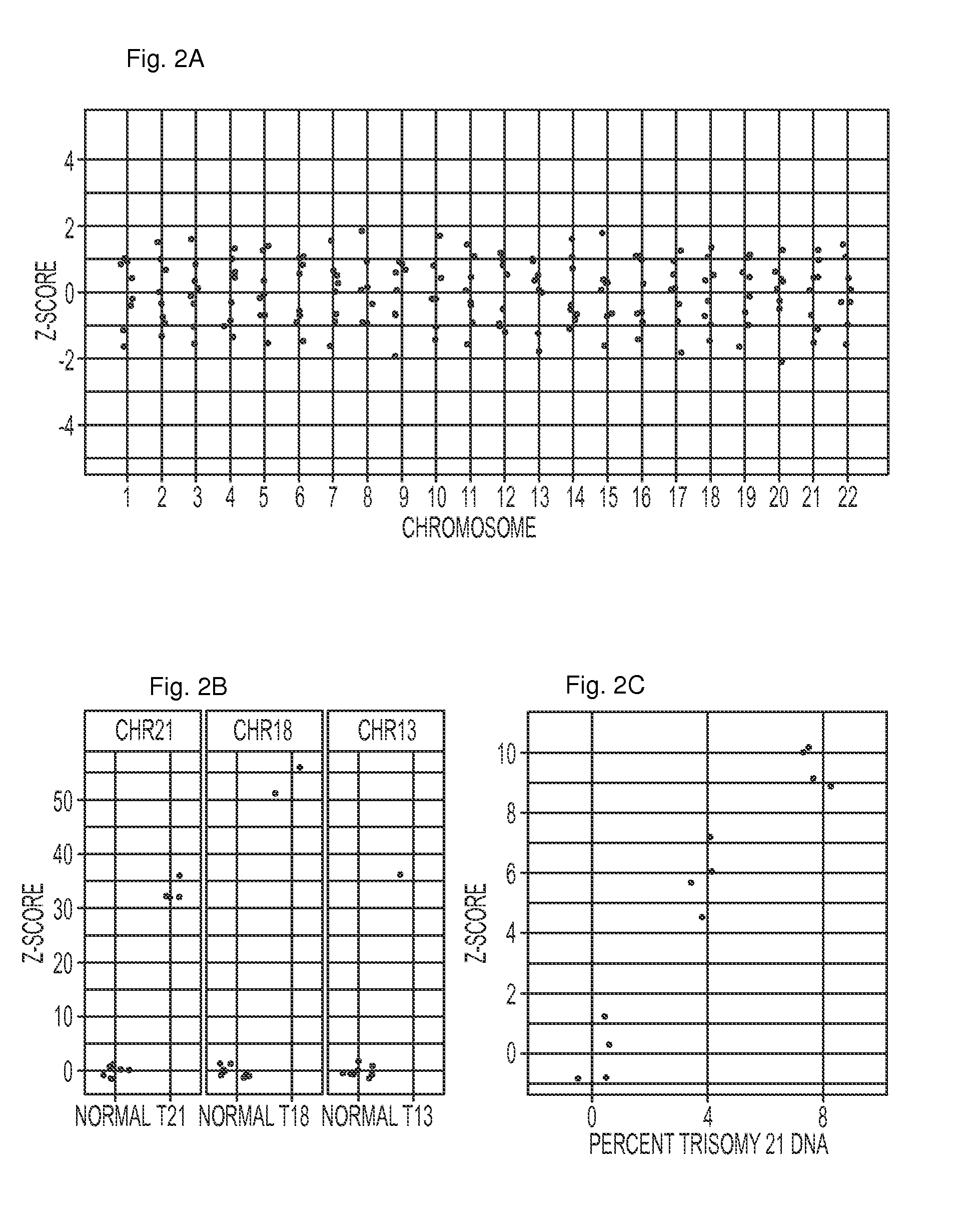
[0059]We next studied the reproducibility of chromosome representation in two additional experiments employing different types of samples, different instruments, and different depths of sequencing. In the first experiment, we analyzed DNA from peripheral blood white blood cells (WBCs) from the same seven individuals who contributed plasma. Four samples were sequenced on a single lane of an Illumina HiSeq 2000, yielding a mean of 10,835,559 uniquely aligned tags per sample (range: 4,905,067 to 16,015,347 tags). The maximum z-scores for any of the samples were 1.0, 1.2, and 1.6 for chromosomes 21, 18, and 13, respectively (Supplementary FIG. 1a).
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| relative frequency | aaaaa | aaaaa |
| area | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com