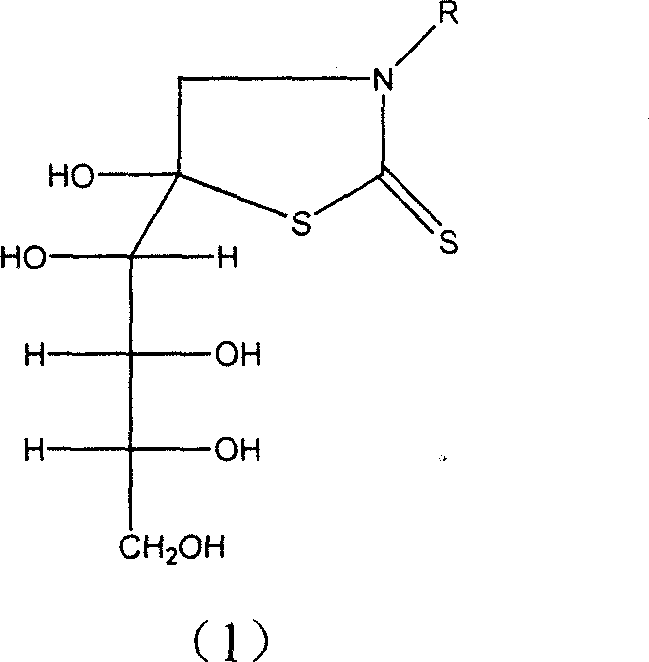
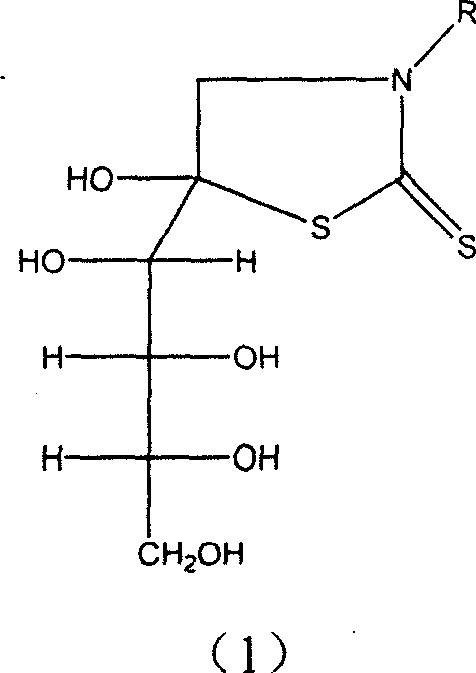
Glucose derivative complex marked with 99mTc, 188 Re or 186Re and its prepn process
A glucose derivative, 99mtc technology, applied in the field of glucose derivative complexes, can solve problems such as low equipment penetration rate, high FDG cost, and difficulty in popularizing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 11
[0105] Example 1.1: [ 99m TcO] 3+ Nuclear labeling compound 1
[0106] Add 0.5mL water and 0.5mL ethanol to 2mg compound 1 to make a solution, add 0.3mL (10mCi / ml) freshly rinsed Na 99m TCO 4 , quickly add 0.2mlSnCl 2 Solution (1mg / mL, 0.1NHCl), adjust pH=8.0~8.5, react at room temperature for 20min, filter with 0.2μm filter membrane, monitor by TLC (acetonitrile / polyamide; normal saline / silica gel), calculate the labeling rate, the labeling rate is 99.26 %, HPLC monitoring (HPLC condition: A phase 0.1% trifluoroacetic acid aqueous solution; B phase 0.1% trifluoroacetic acid acetonitrile solution; Gradient: 0~10min: A=100%, B=0; 10~20min: A=50 %, B=50%; 20-30min: A=0, B=100%). HPLC retention time is 17.20min, 99.94%.
Embodiment 12
[0107] Example 1.2: [ 99m TcO] 3+ Nuclear labeling compound 3
[0108] Add 0.5mL water and 0.5mL ethanol to 2mg compound 3 to make a solution, add 0.3mL (10mCi / mL) freshly rinsed Na 99m TCO 4 , quickly add 0.2mlSnCl 2 Solution (1mg / mL, 0.1NHCl), adjust pH=8.0~8.5, react at room temperature for 20min, filter with 0.2μm filter membrane, monitor by TLC (acetonitrile / polyamide; normal saline / silica gel), calculate the labeling rate, the labeling rate is greater than 99%. HPLC monitoring (HPLC conditions: A phase 0.1% trifluoroacetic acid aqueous solution; B phase 0.1% trifluoroacetic acid acetonitrile solution; Gradient: 0~10min: A=100%, B=0; 10~20min: A=50%, B=50%; 20-30min: A=0, B=100%). HPLC retention time is 16.60min, 99.93%.
Embodiment 13
[0110] [ 99m TcO] 3+ Nuclear labeling compound 6
[0111] Add 0.5mL water and 0.5mL ethanol to 2~3mg compound 6 to make a solution, add 0.3mL (10mCi / mL) freshly rinsed Na 99m TCO 4 , quickly add 0.2mlSnCl 2 Solution (1mg / mL, 0.1NHCl), adjust pH=8.0~8.5, react at room temperature for 20min, filter with 0.2μm filter membrane, monitor by TLC (acetonitrile / polyamide; normal saline / silica gel), calculate the labeling rate, the labeling rate is greater than 99%. HPLC monitoring (HPLC conditions: A phase 0.1% trifluoroacetic acid aqueous solution; B phase 0.1% trifluoroacetic acid acetonitrile solution; Gradient: 0~10min: A=100%, B=0; 10~20min: A=50%, B=50%; 20-30min: A=0, B=100%). HPLC retention time is 16.70min, 86.0
[0112] [ 99m TcO] 3+ Nuclear labeling compound 2
[0113] Add 0.5mL water and 0.5mL ethanol to 2mg compound 2 to make a solution, add 0.3mL (10mCi / ml) freshly rinsed Na 99m TCO4 , quickly add 0.2mlSnCl 2 Solution (1mg / mL, 0.1NHCl), adjust pH=8.0~8.5, reac...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com