Method and reagent case for detecting hepatitis b virus anterior C/BCP region gene mutation
A technology of hepatitis B virus and kit, applied in reverse dot hybridization technology to quickly detect mutations in the pre-C/BCP region of HBV gene, and reverse dot hybridization technology to quickly detect mutations in the pre-C region and BCP region of HBV gene, which can solve the problem of detecting genes The mutation operation is cumbersome, the mutation form of the specific site cannot be judged, and the promotion and application are limited.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0082] Embodiment 1: Preparation of sample target nucleic acid
[0083] Use a disposable sterile syringe to extract 2 ml of venous blood from the subject, inject it into a sterile dry glass tube, and place it at room temperature (22-25°C) for 30-60 minutes. Centrifuge at 1500rpm for 5 minutes; absorb 100μl of serum from the upper layer and transfer to a 1.5ml sterilized centrifuge tube. Then add an equal volume of concentrated DNA solution into the centrifuge tube and mix well, centrifuge at 12000rpm for 10 minutes, and collect the precipitate at the bottom of the tube. Add 50 μl of conventional DNA extraction solution to the precipitate, mix well, bathe in boiling water for 10 minutes and centrifuge at 12000 rpm for 10 minutes. Take 2 μl of the supernatant as a template for the PCR reaction. In the formulation of the concentrated solution, we have used different concentrations of polyethylene glycol (3% to 10%) and sodium chloride in combination, and 3% to 10% of polyethyle...
Embodiment 2
[0084] Embodiment 2: PCR amplification of target nucleic acid
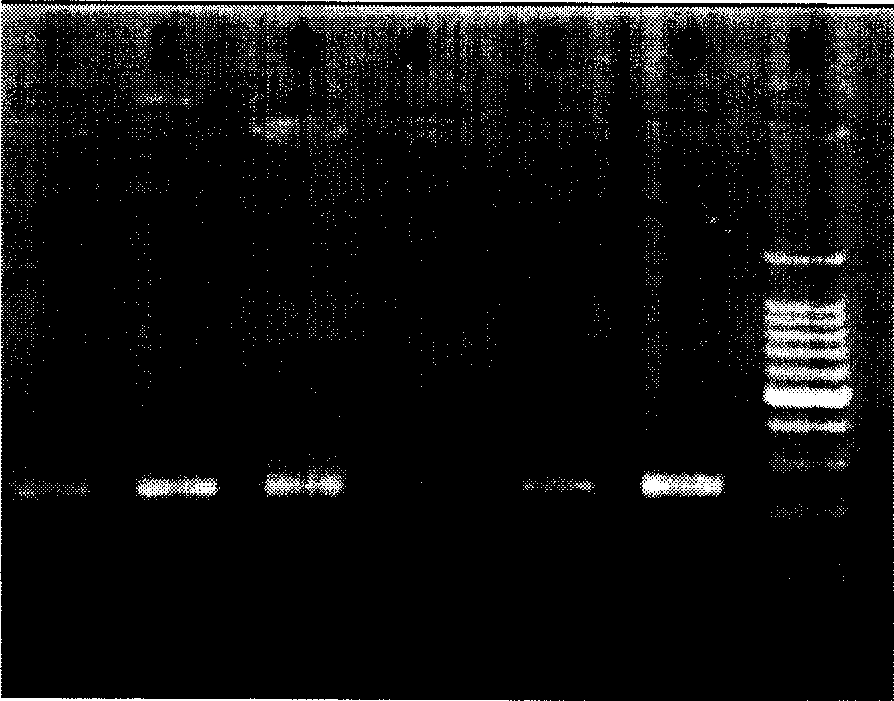
[0085] Take several tubes of PCR reaction solution for a single tube, add 2 μl template (or negative and positive standards) directly, mix well, centrifuge briefly (3 seconds), and put each reaction tube into the PCR machine. After pretreatment at 50°C for 3 minutes, amplify according to the following conditions: 93°C for 30 seconds, 55°C for 45 seconds, 72°C for 30 seconds, 40 cycles, and finally 72°C for 7 minutes. The amplified products were detected by 2% agarose gel electrophoresis (see attached figure 2 ).
Embodiment 3
[0086] Example 3: Sample reverse dot blot detection of six known genotypes
[0087] Before hybridization, hybridization solution I (2×SSC-0.15% SDS) was mixed with hybridization solution II and preheated to 42°C for use. According to the number of samples to be tested, take a corresponding number of 1.5ml centrifuge tubes, add 0.5ml hybridization solution I to each tube and preheat to 42°C. After the PCR amplification product was denatured at 98°C for 8 minutes, it was placed in an ice-water mixture for 8-10 minutes. Then take 1000μl hybridization solution I+2μl solution I (1000:2) mixed solution as the binding solution and store it at 4°C for later use, and take solution II: solution III: solution IV (1900:200:1) mixture (19ml solution II+2ml Solution III + 10 μl solution IV) was used as a chromogenic solution protected from light for later use.
[0088] Before hybridization, fill the reaction chamber with distilled water, place the metal perforated plate, and turn on the w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com