Process for detecting DNA methylation of paraffin specimen
A methylation and specimen technology, applied in the field of medical biology detection, can solve the problems of easy random degradation, troublesome operation, unstable results, etc., and achieve the effects of being beneficial to environmental protection, saving time, and avoiding repeated precipitation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
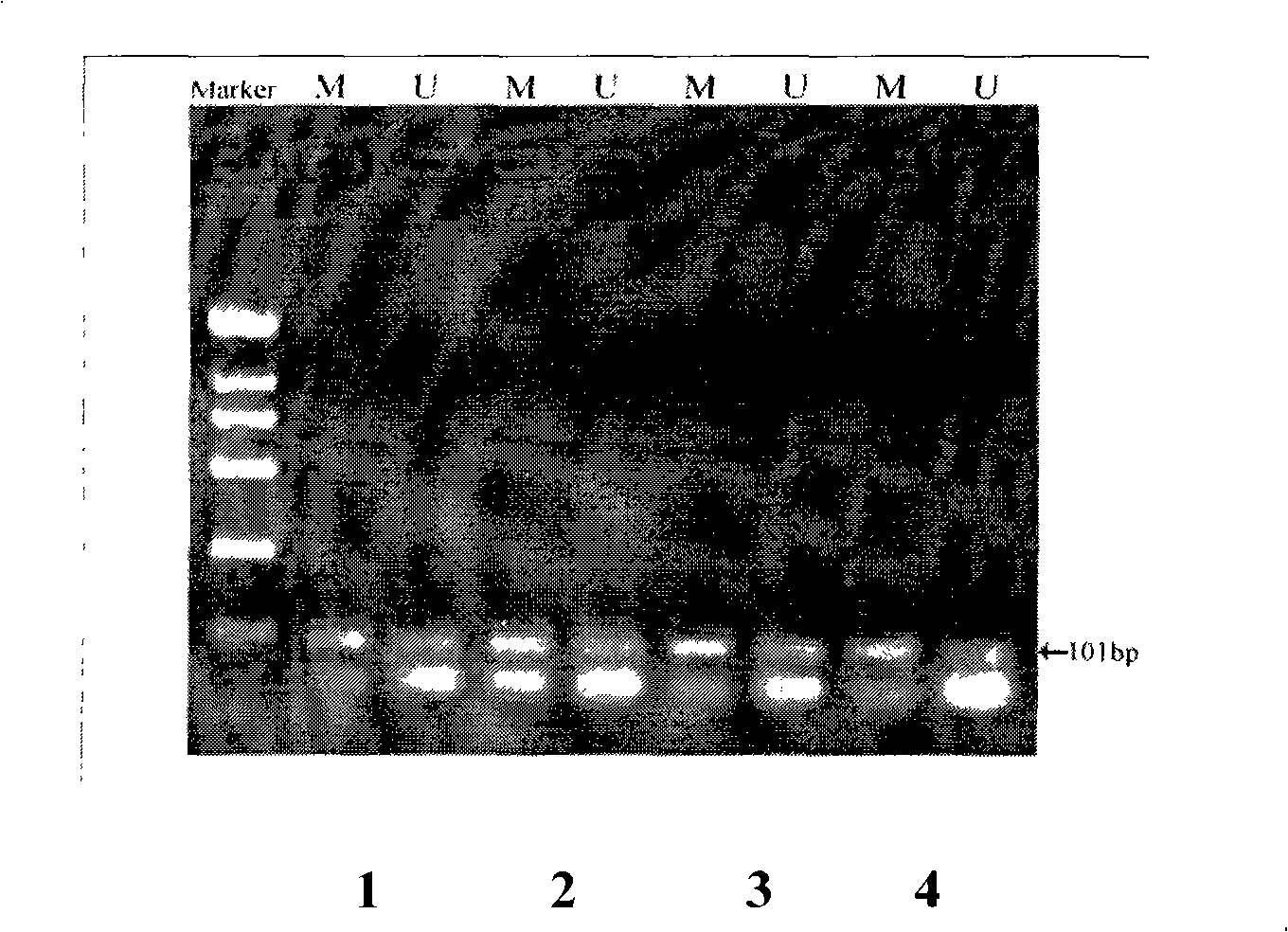
[0060] Example 1. Detection of methylation experiment of GSTP1 gene in colon cancer, gastric cancer, liver cancer, ovarian cancer paraffin specimens
[0061] According to literature reports, the GSTP1 gene in patients with colon cancer, gastric cancer, liver cancer, and ovarian cancer is often in a methylated state (Nakayama M, Bennett CJ, Hicks JL, et al.Hypermethylation of the human glutathione S-transferase-k gene (GSTP1) CpG island is present in a subset of proliferative inflammatory atrophylesions but not in normal or hyperplastic epithelium of the prostate: adetailed study using laser-capture microdissection. Am J Pathol 2003; 163: 923-33).
[0062] The paraffin specimens of sigmoid colon carcinoma (030081D), gastric angle adenocarcinoma grade III (030090B), antral adenocarcinoma (030083C), and ovarian cancer (030082C) were obtained from the Department of Pathology, Dongfang Hepatobiliary Hospital, and the experimental methods for each specimen were the same.
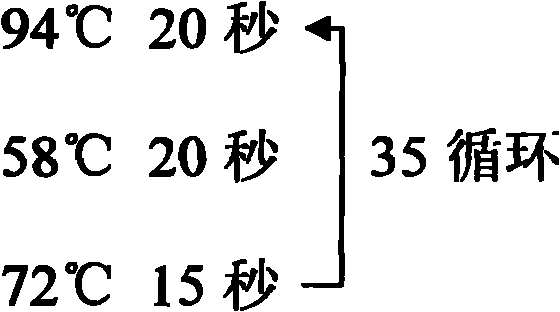
[0063] Th...
Embodiment 2
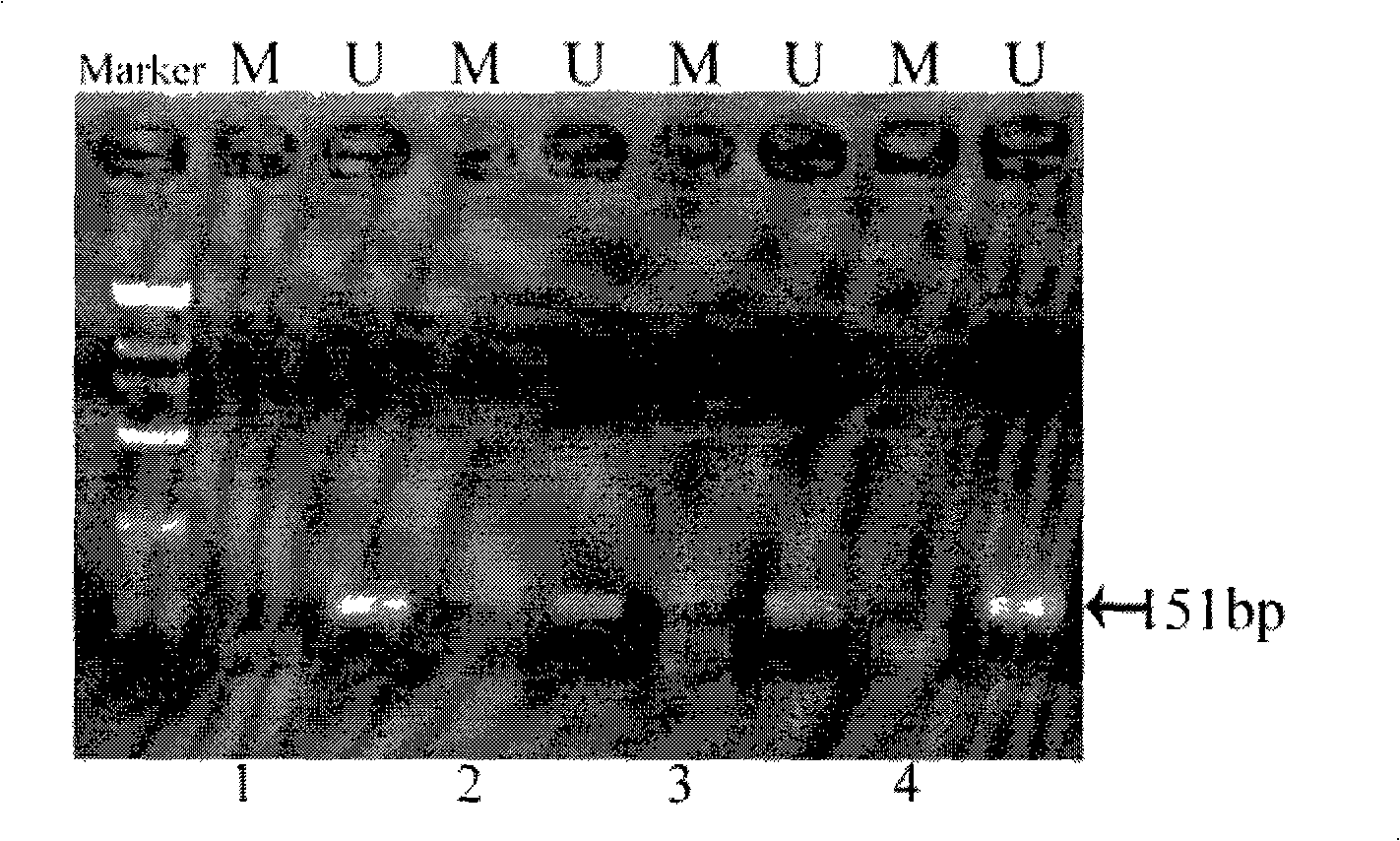
[0105] Example 2. Detection of p16 gene methylation experiment in paraffin wax specimens of colon cancer, gastric cancer, liver cancer and ovarian cancer
[0106] The samples of sigmoid colon cancer (030081D), gastric angle adenocarcinoma grade III (030090B), gastric antrum adenocarcinoma (030083C), and ovarian cancer (030082C) were the same as in Example 1.
[0107] The operation steps are as follows:
[0108] 1. Dewaxing:
[0109] 1. The paraffin specimen slices were fixed on glass slides: the thickness of the slices was 5 μm, and two slices were taken from each slide; baked at 68°C for 1 hour;
[0110] 2. Place the glass slide on the slide rack, immerse in xylene for dewaxing twice, 10 minutes each time.
[0111] 2. Specimen hydration:
[0112] The slides were immersed in absolute ethanol, 95% ethanol, 85% ethanol, and 70% ethanol in sequence, for 10 minutes each time, and finally immersed in distilled water to wash twice, 10 minutes each time.
[0113] 3. DNA sodium bi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap