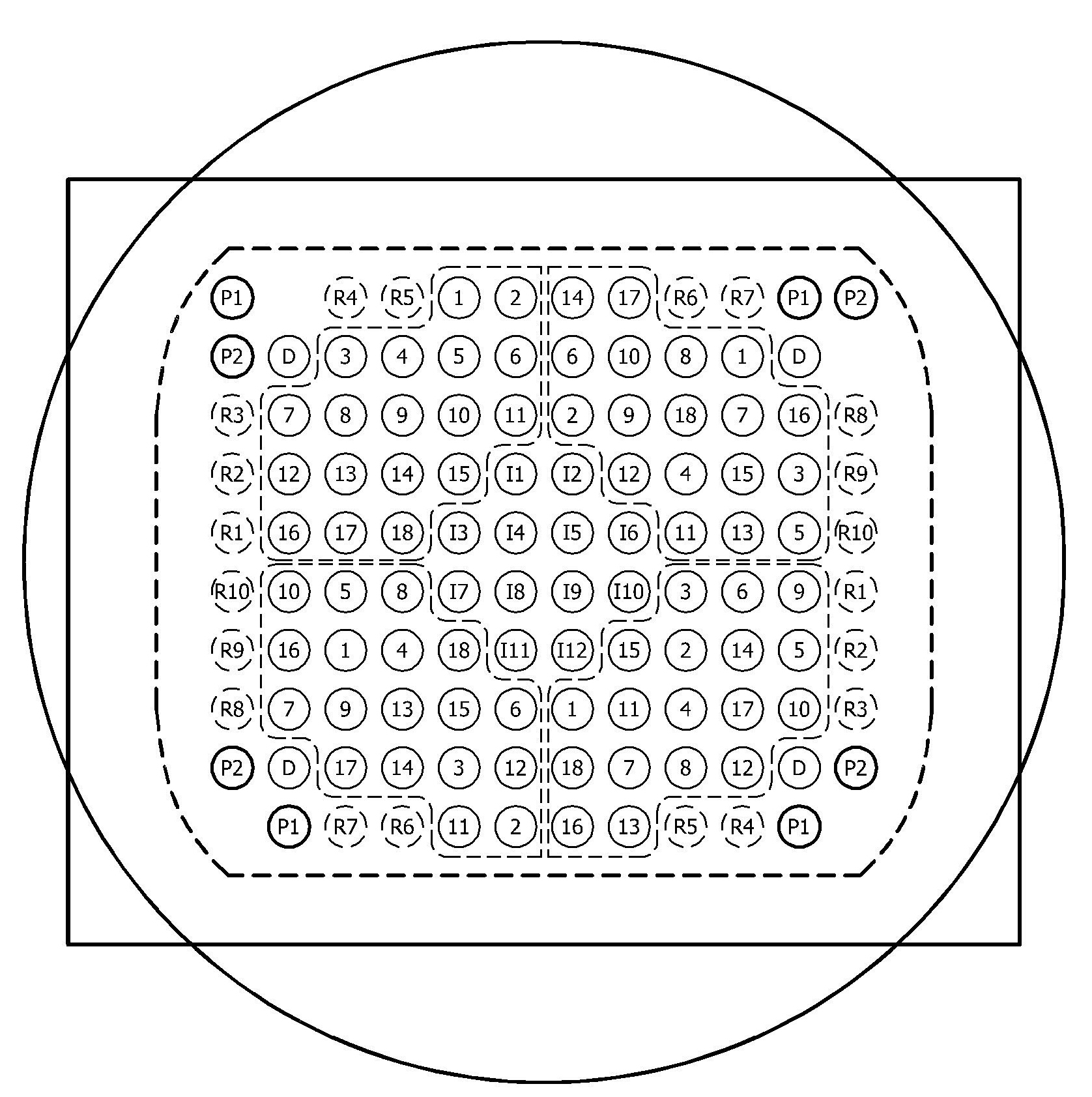
Method for immobilizing nucleic acids on a support
一种核酸、载体的技术,应用在生物化学设备和方法、微生物的测定/检验等方向,能够解决危害应用、影响分子与互补序列杂交的能力、核酸分子损伤等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
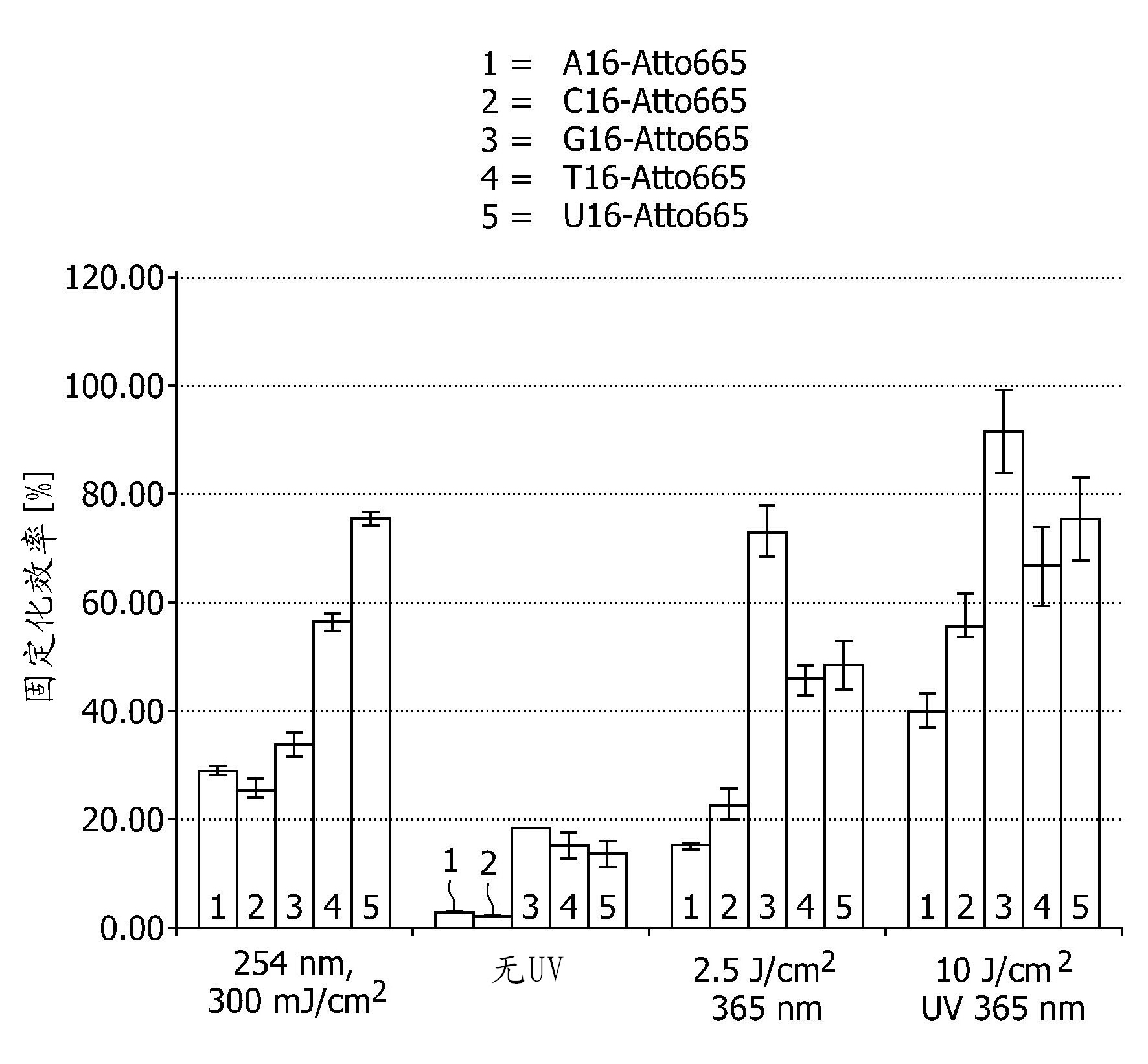
[0136] Example 1 - Test of Immobilization Efficiency at Different Wavelengths
[0137] The immobilization efficiency of different immobilization methods was tested on Nytran SPC nylon membrane. Specifically, light at a wavelength of 254 nm, non-ultraviolet light, and light at a wavelength of 365 nm were used for exposure of nucleic acid molecules in exposure periods of 5 seconds or 20 seconds of exposure. As nucleic acid molecules, different oligonucleotides were used, comprising 16 nucleotides of only one base (ie 16A, 16C, 16G, 16T, 16U), each 16-mer labeled with Atto655 dye.
[0138] The parameter tested was the nucleic acid immobilization efficiency or recovery, i.e. the ratio between immobilized and precipitated oligonucleotides, corrected for the loss of fluorophore density during UV application. After UV application, samples were washed to remove unbound material. As washing buffer, 5x SSC containing 0.1% SDS and 0.1 mg / ml herring sperm DNA was used. Washing / blocking...
Embodiment 2
[0142] Example 2 - Recovery Test and Sensitivity Test of T-Tail-Containing Nucleic Acids
[0143] Sensitivity, i.e. the amount of analyte captured per unit time, was tested in a real-time hybridization assay. Nytran N or Nytran SPC nylon films were used for experiments.
[0144] The assay is performed using capture oligonucleotides (i.e. immobilized precipitated nucleic acid molecules) that either do not contain a T tail or contain a T16 tail, a stretch of 16 thymines. These experiments were performed in a flow cell, which is a device in which a membrane is clamped and hybridization fluid is pumped through the membrane. Figure 3A , the number of cycles is depicted on the x-axis, which is equal to time (1 cycle takes 1 minute). Hybridization is accomplished with complementary DNA. The hybridization buffer is 5×SSC, 0.1% SDS, 0.1mg / ml herring sperm DNA. The temperature was set at 50°C.
[0145] As can be obtained from Figure 3A, oligonucleotides containing the T16 tail sho...
Embodiment 3
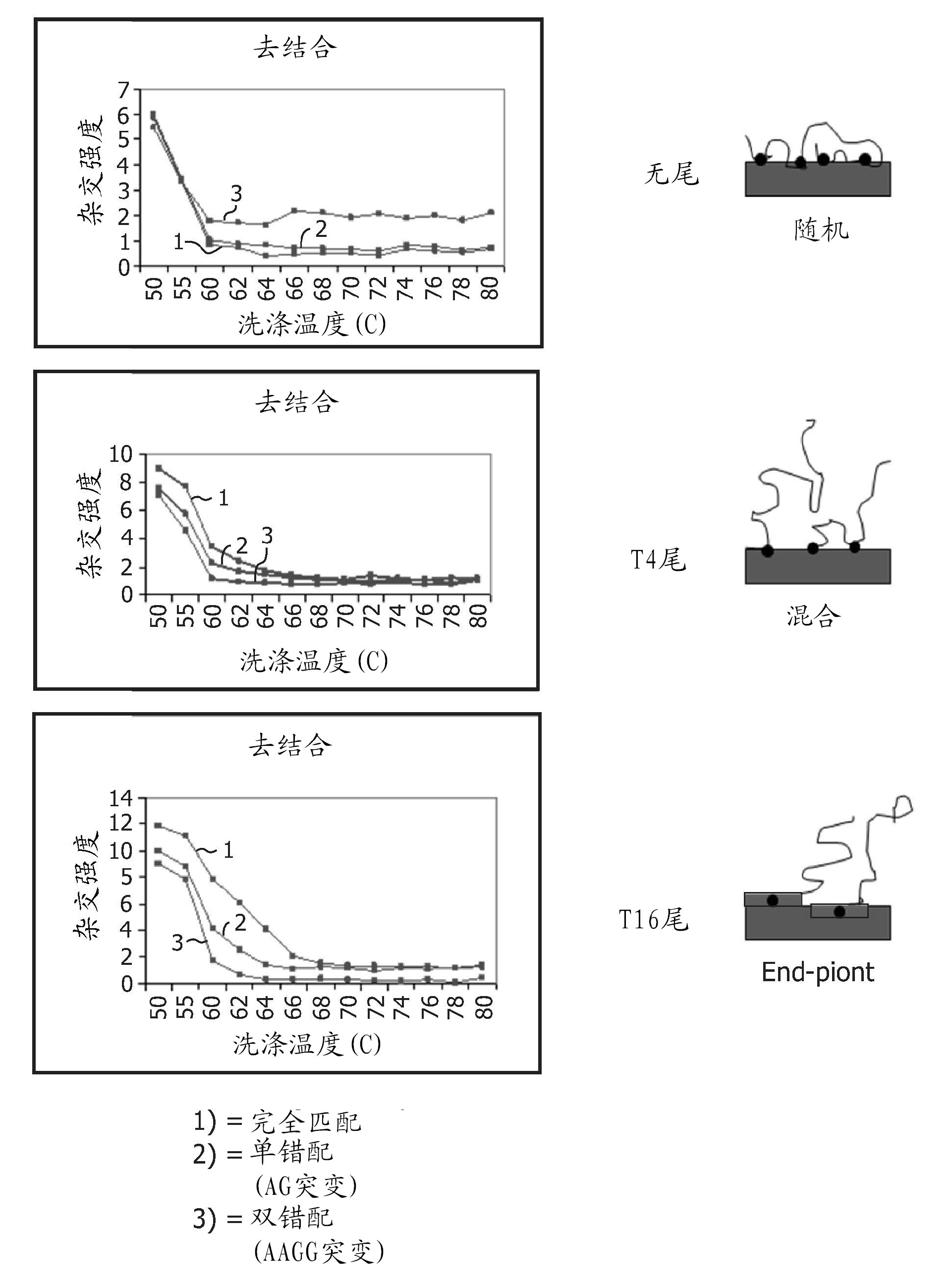
[0148] Example 3—Specificity Test of Immobilized Nucleic Acids
[0149] The specificity of the immobilized nucleic acids, ie the ability to distinguish between matched and unmatched targets, was tested in a binding assay.
[0150] The assay is performed with immobilized nucleic acids (capture probes) containing 0, 4 or 16 Ts. Different capture probes were used, including perfect matches, single mismatches ((AG) mutations) and double mismatches ((AAGG) mutations). PCR products were used for hybridization. Hybridization is performed in a flow cell, which is a device in which a membrane is clamped and hybridization fluid is pumped through the membrane. The temperature was set at 50°C. Hybridization was performed for 1 hour. After hybridization, the buffer was changed to 2 × SSC, and the temperature was increased at 1°C / min to generate melting curves and assess specificity.
[0151] Figure 4 depicts the debinding curves for complementary, single-mismatched and double-mismatch...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com