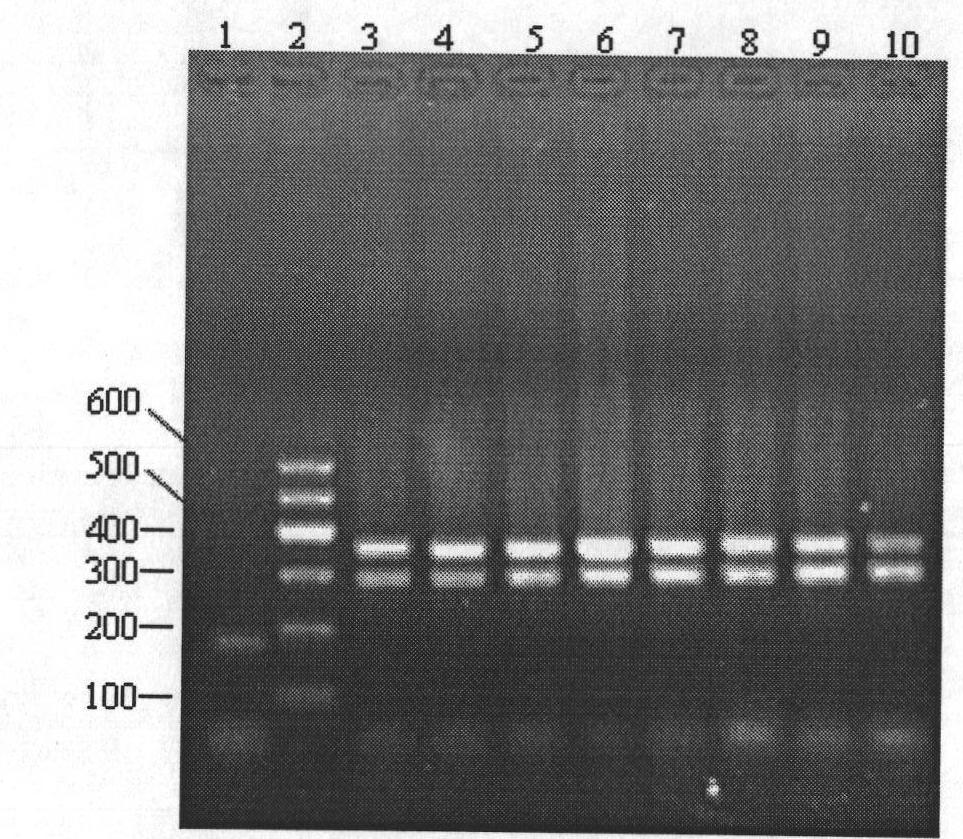
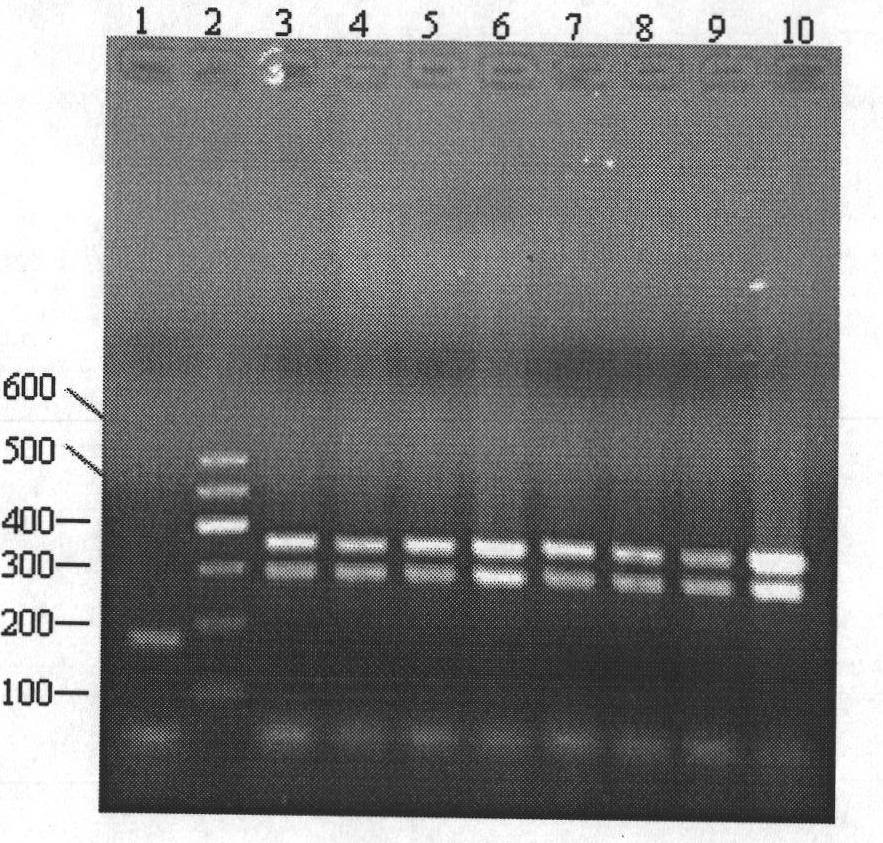
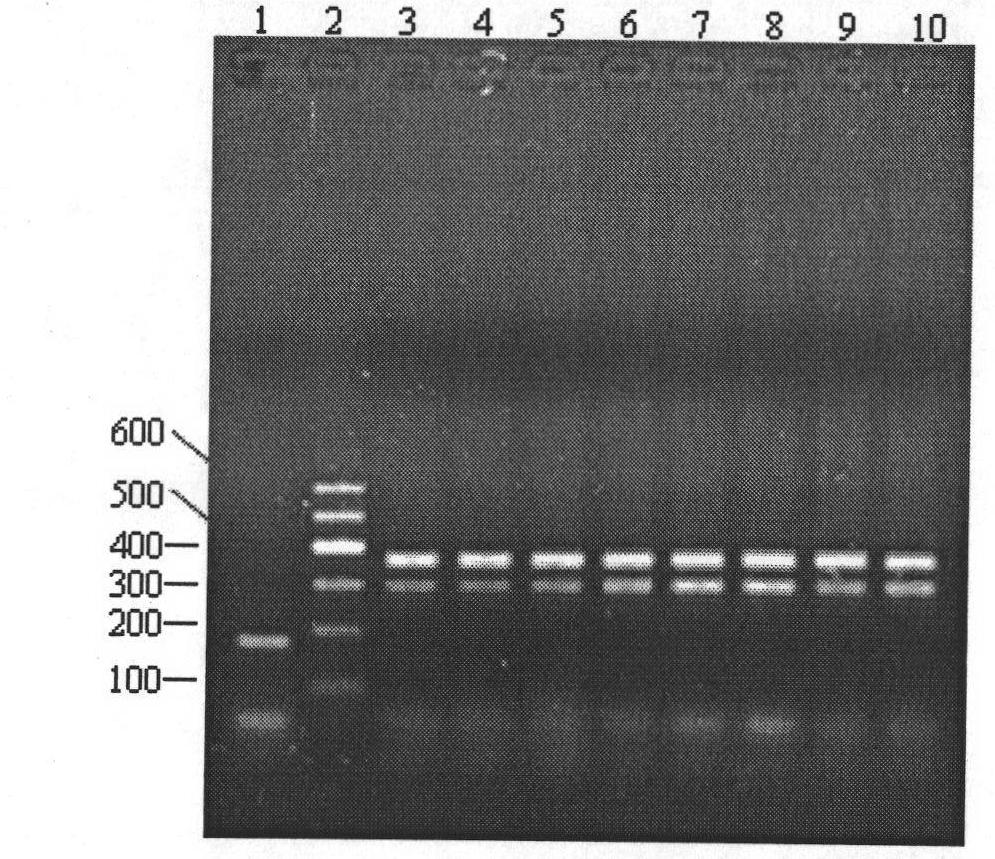
Method for identifying gynostemma pentaphylla and making distinction between gynostemma pentaphylla and cayratia japonica at deoxyribonucleic acid (DNA) level
A technology of Gynostemma pentaphyllum and black twig, applied in the field of identifying Gynostemma pentaphyllum at the DNA level and distinguishing it from black twig
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0057] 1. Extract DNA:
[0058] DNAs of Gynostemma cucurbitaceae and Ussica berry of grape family were extracted respectively by the CTAB method improved by the inventor.
[0059] (1) DNA extraction from fresh samples of Cucurbitaceae Gynostemma
[0060] ① Weigh 0.5g leaves and place them in a pre-cooled mortar, add liquid nitrogen and grind them into fine powder, quickly transfer them to a 7ml centrifuge tube, add 3ml preheated 1.5×CTAB extraction buffer, mix well, and place in a 65°C water bath Keep warm for 20min.
[0061] ② Cool the extract from step ① to room temperature, and centrifuge at 7500rpm for 10min. Take the supernatant, add an equal volume of chloroform / isoamyl alcohol (24 / 1) for extraction, mix well, and centrifuge at 7500rpm for 10min.
[0062] ③ Take the centrifuged supernatant in step ②, add 1 / 10 volume of 1×CTAB extraction buffer and an equal volume of chloroform / isoamyl alcohol (24 / 1), mix well, and centrifuge at 7500rpm for 10min.
[0063] ④ Take the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com