Specific sequence characterized amplified region (SCAR) marker and rapid polymerase chain reaction (PCR) detection method for Heterodera filipjevi
A technology for the detection of Phillips spores, which is applied in the field of rapid PCR detection of Phillips cyst nematode SACR specificity, and can solve the problems of reporting and lack of specific primers for Phillips cysts nematode
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1: DNA fragments, specific primers for Phillips cyst nematode specific SCAR markers
[0046] 1.1 Extraction of DNA from Philip's cyst nematode
[0047] Pick one cyst from each group of Phillips cyst nematode, cereal cyst nematode, and soybean cyst nematode by hand, put it into 10 μl sterilized redistilled water, and freeze it in a -20°C refrigerator for 1 hour. A glass rod sterilized with 75% alcohol was rotated in an eppendorf tube until the ice melted, 7 μl of 10X PCR buffer and 3 μl of proteinase K solution (600 μg / ml) were added, and then frozen at -20°C for at least 2 hours. After 2 hours, take the eppendorf tube out of the refrigerator and incubate at 65°C for 1.5h; then denature proteinase K at 95°C for 10 minutes, centrifuge at 1000 rpm for 1 minute, and take the supernatant DNA suspension and store it at -20°C for later use.
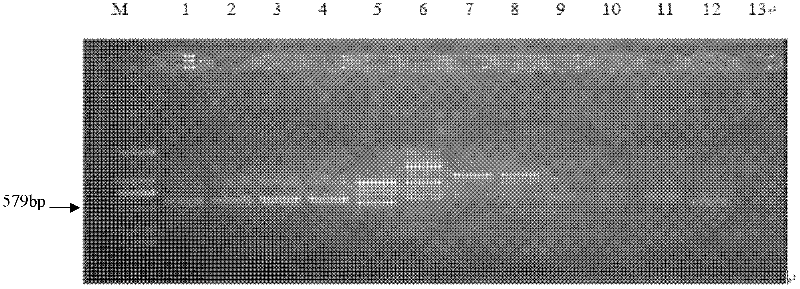
[0048] 1.2 Screening of Phillips cyst nematode-specific RAPD markers (SCAR markers)
[0049] Drawing on the results of genetic...
Embodiment 2
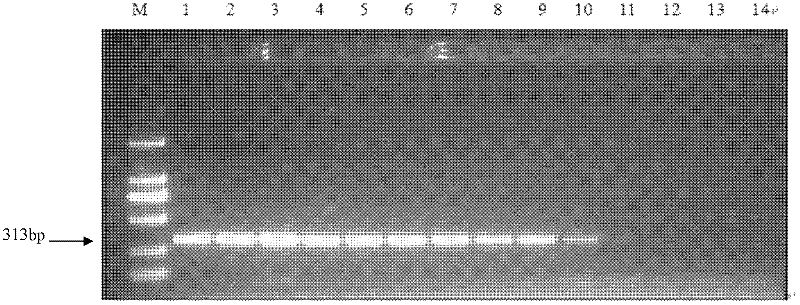
[0060] Example 2: Molecular detection of Phillips cyst nematode-specific SCAR marker primers
[0061]Phillips cyst nematode-specific SCAR marker primers HfF1 and HfR1 constructed in the present invention were synthesized by Shanghai Sangon Biology Co., Ltd. The total volume of the specific SCAR marker amplification reaction system was 50 μl, which contained 5 μl 10XPCR buffer (containing Mg++); 4 μl 2.5mM dNTP; 1.0μl HfF1 (0.1μg / μl); 1.0μl HfR1 (0.1μg / μl); 1U Taq DNA polymerase (5U / μl); 2μl template DNA (Philips cyst nematode and other cyst nematode population extraction DNA); ddH 2 O 36.5 μl.
[0062] The applied Phillips cyst nematode-specific SCAR marker primer sequences are as follows:
[0063] HfF1: 5'-AATCCTTACACCGTTGTTTATTA-3'
[0064] HfR1: 5'-ATTCTTCCTCCTTCTCCCACTAT-3'
[0065] The PCR amplification program was as follows: pre-denaturation at 94°C for 4 min, followed by 35 cycles of 94°C for 30S, 56°C for 30S, 72°C for 1 min, 72°C for 10 min, and storage at 4°C. ...
Embodiment 3
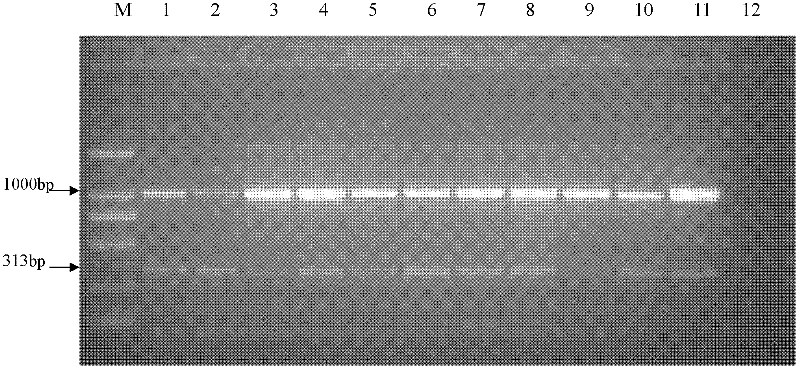
[0067] Embodiment 3: One-step double PCR method of specific SCAR marker primers and universal primers (TW81 and AB28) detects Phillips cyst nematode
[0068] In the present invention, the constructed philip cyst nematode specific SCAR marker primers HfF1 and HfR1 and the universal primers TW81 and AB28 are placed in the same PCR reaction, and HfF1 and HfR1 specifically amplify the SCAR of philip cyst nematode genomic DNA The marker fragment, primers TW81 and AB28 were used to amplify the DNA fragment of the rDNA gene ITS extension region. The one-step double PCR method adopted in the present invention is the world's first molecular diagnosis method of Philip's cyst nematode, and the molecular detection method contains two fragments of genomic DNA and ribosomal DNA.
[0069] The molecular detection method of Phillips cyst nematode of the present invention contains a 25 μl PCR reaction system, and the proportion is as follows: 10X PCR-buffer (containing Mg++) 2.5 μl, dNTPs 2 μl,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com