Sample analysis method and assay kit for use in the method
A detection kit and analysis method technology, applied in the field of nucleic acid analysis, can solve the problems of increased chip time detection cost, large number of samples, etc., and achieve the effects of rapid analysis, reduced inspection cost, and simple analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0045] Hereinafter, embodiments of the present invention will be described.
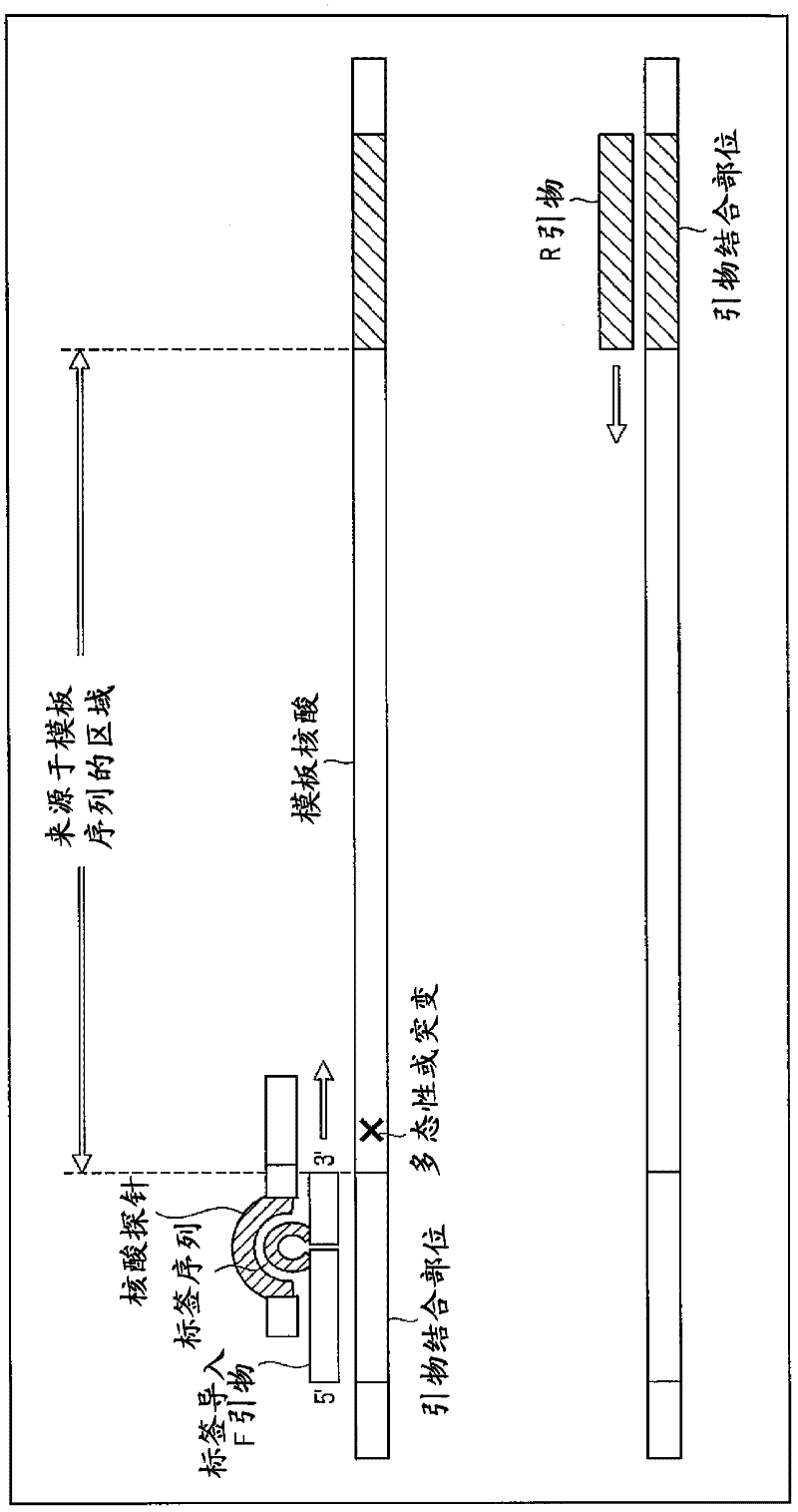
[0046] figure 1 is a diagram showing tag sequence introduction primers and nucleic acid probes.
[0047]
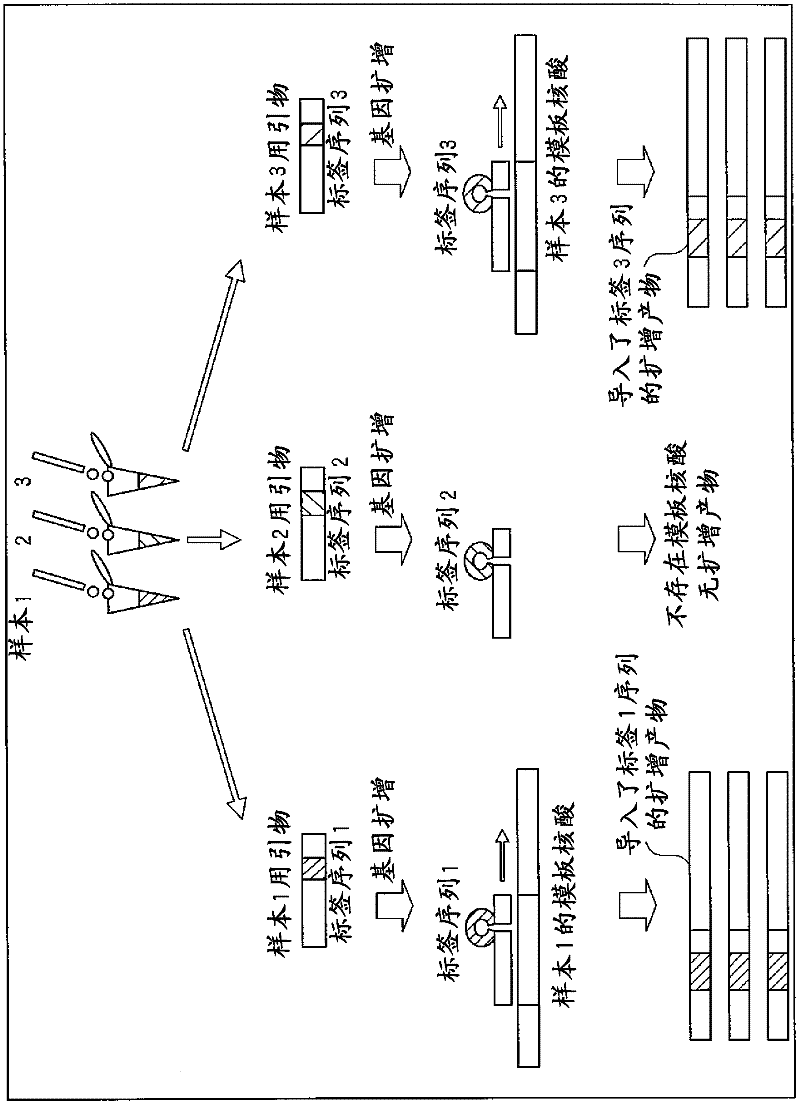
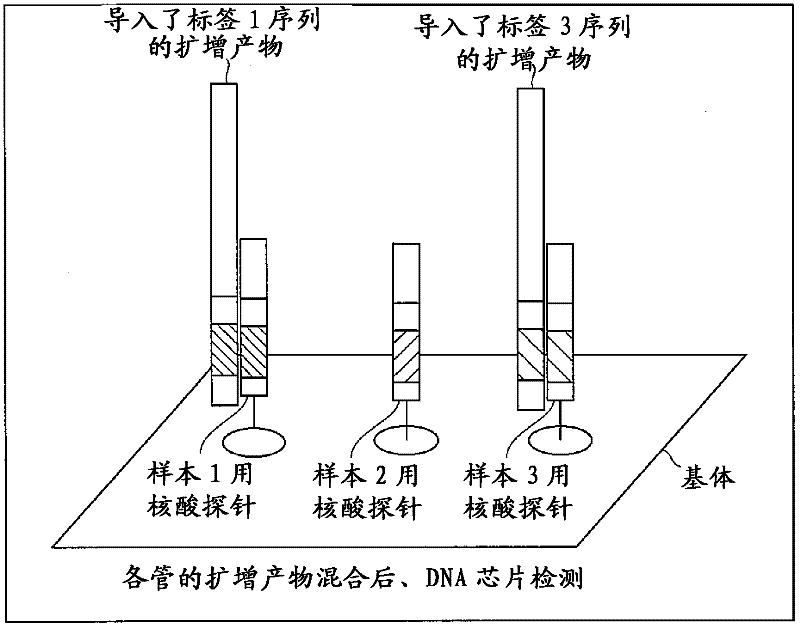
[0048] Such as figure 1 A sequence that binds to the primer-binding site of the template nucleic acid and a tag sequence for analyzing multiple samples are introduced into the primer as exemplified by the tag-introducing F primer.
[0049] A different sequence is used for the tag sequence depending on the sample. For example, when a tag sequence of 5 bases is prepared and the tag sequence is composed of 4 types of bases, A, T, C, and G, there are 45, that is, 1024 types of tag sequences as candidates. However, since there is a high risk of mismatch hybridization when using tag sequences different in one base, it is preferable to prepare tag sequences different in sequence by two or more bases. As for the number of label sequences to be prepared, it is only necessary to prepare the number o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com